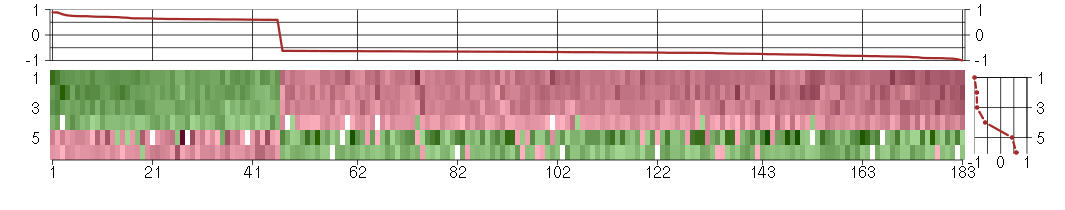

Under-expression is coded with green,
over-expression with red color.


membrane
Double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
cell
The basic structural and functional unit of all organisms. Includes the plasma membrane and any external encapsulating structures such as the cell wall and cell envelope.
intrinsic to membrane
Located in a membrane such that some covalently attached portion of the gene product, for example part of a peptide sequence or some other covalently attached moiety such as a GPI anchor, spans or is embedded in one or both leaflets of the membrane.
membrane part
Any constituent part of a membrane, a double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
all
This term is the most general term possible
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
membrane part
Any constituent part of a membrane, a double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.

ABCE1ATP-binding cassette, sub-family E (OABP), member 1 (201873_s_at), score: -0.7 ACSL1acyl-CoA synthetase long-chain family member 1 (201963_at), score: -0.68 ACSL4acyl-CoA synthetase long-chain family member 4 (202422_s_at), score: -0.84 ACTR3BARP3 actin-related protein 3 homolog B (yeast) (218868_at), score: -0.84 ADARadenosine deaminase, RNA-specific (201786_s_at), score: 0.65 ADORA2Badenosine A2b receptor (205891_at), score: 0.74 AHI1Abelson helper integration site 1 (221569_at), score: -0.83 AIM1absent in melanoma 1 (212543_at), score: 0.7 AKAP9A kinase (PRKA) anchor protein (yotiao) 9 (210962_s_at), score: -0.65 ANXA10annexin A10 (210143_at), score: -0.69 APAF1apoptotic peptidase activating factor 1 (204859_s_at), score: -0.68 ARFGAP3ADP-ribosylation factor GTPase activating protein 3 (202211_at), score: -0.63 ATG4AATG4 autophagy related 4 homolog A (S. cerevisiae) (213115_at), score: -0.65 ATP2C1ATPase, Ca++ transporting, type 2C, member 1 (211137_s_at), score: -0.69 AZI25-azacytidine induced 2 (218043_s_at), score: -0.66 BMP6bone morphogenetic protein 6 (206176_at), score: -0.75 BRWD1bromodomain and WD repeat domain containing 1 (214820_at), score: -0.7 C12orf29chromosome 12 open reading frame 29 (213701_at), score: -0.63 C14orf45chromosome 14 open reading frame 45 (220173_at), score: -0.76 C14orf94chromosome 14 open reading frame 94 (218383_at), score: 0.72 C15orf33chromosome 15 open reading frame 33 (216411_s_at), score: -0.72 C4orf43chromosome 4 open reading frame 43 (218513_at), score: -0.87 C5orf44chromosome 5 open reading frame 44 (218674_at), score: -0.83 C7orf64chromosome 7 open reading frame 64 (221596_s_at), score: -0.65 CALB2calbindin 2 (205428_s_at), score: -0.77 CAPZA2capping protein (actin filament) muscle Z-line, alpha 2 (201237_at), score: -0.63 CASC3cancer susceptibility candidate 3 (207842_s_at), score: 0.61 CCDC76coiled-coil domain containing 76 (219130_at), score: -0.77 CD2APCD2-associated protein (203593_at), score: -0.66 CDC25Bcell division cycle 25 homolog B (S. pombe) (201853_s_at), score: 0.7 CDC2L6cell division cycle 2-like 6 (CDK8-like) (212899_at), score: 0.6 CDC42EP4CDC42 effector protein (Rho GTPase binding) 4 (214721_x_at), score: 0.74 CDH6cadherin 6, type 2, K-cadherin (fetal kidney) (205532_s_at), score: -0.69 CHORDC1cysteine and histidine-rich domain (CHORD)-containing 1 (218566_s_at), score: -0.75 CHUKconserved helix-loop-helix ubiquitous kinase (209666_s_at), score: -0.69 CICcapicua homolog (Drosophila) (212784_at), score: 0.59 CITcitron (rho-interacting, serine/threonine kinase 21) (212801_at), score: 0.63 CMPK1cytidine monophosphate (UMP-CMP) kinase 1, cytosolic (217870_s_at), score: -0.74 CRTC3CREB regulated transcription coactivator 3 (218648_at), score: 0.89 CScitrate synthase (208660_at), score: 0.65 CSGALNACT2chondroitin sulfate N-acetylgalactosaminyltransferase 2 (222235_s_at), score: -0.74 CTAGE5CTAGE family, member 5 (215930_s_at), score: -0.65 CXCL6chemokine (C-X-C motif) ligand 6 (granulocyte chemotactic protein 2) (206336_at), score: -0.67 DENND4CDENN/MADD domain containing 4C (205684_s_at), score: -0.64 DENND5BDENN/MADD domain containing 5B (215058_at), score: -0.85 DIS3DIS3 mitotic control homolog (S. cerevisiae) (218362_s_at), score: -0.66 DNAJB9DnaJ (Hsp40) homolog, subfamily B, member 9 (202843_at), score: -0.93 DNAJC6DnaJ (Hsp40) homolog, subfamily C, member 6 (204720_s_at), score: -0.69 DOPEY1dopey family member 1 (40612_at), score: -0.79 EAF2ELL associated factor 2 (219551_at), score: -0.95 EGFepidermal growth factor (beta-urogastrone) (206254_at), score: -0.7 EIF2AK3eukaryotic translation initiation factor 2-alpha kinase 3 (218696_at), score: -0.7 EIF4Eeukaryotic translation initiation factor 4E (201436_at), score: -0.82 ESM1endothelial cell-specific molecule 1 (208394_x_at), score: -0.7 EVI1ecotropic viral integration site 1 (221884_at), score: -0.67 F2RL2coagulation factor II (thrombin) receptor-like 2 (206795_at), score: -0.63 FAM155Afamily with sequence similarity 155, member A (214825_at), score: -0.66 FAM18Bfamily with sequence similarity 18, member B (218446_s_at), score: -0.79 FAM8A1family with sequence similarity 8, member A1 (203420_at), score: -0.68 FAT4FAT tumor suppressor homolog 4 (Drosophila) (219427_at), score: -0.64 FBXL14F-box and leucine-rich repeat protein 14 (213145_at), score: 0.71 FICDFIC domain containing (219910_at), score: -0.68 FLOT1flotillin 1 (210142_x_at), score: 0.61 FOXC2forkhead box C2 (MFH-1, mesenchyme forkhead 1) (214520_at), score: 0.64 FPGTfucose-1-phosphate guanylyltransferase (205140_at), score: -0.66 G3BP2GTPase activating protein (SH3 domain) binding protein 2 (208841_s_at), score: -0.63 GFOD1glucose-fructose oxidoreductase domain containing 1 (219821_s_at), score: -0.8 GHRgrowth hormone receptor (205498_at), score: -0.91 GKglycerol kinase (207387_s_at), score: -0.77 GK3Pglycerol kinase 3 pseudogene (215966_x_at), score: -0.91 GMCL1germ cell-less homolog 1 (Drosophila) (218458_at), score: -0.85 GOLT1Bgolgi transport 1 homolog B (S. cerevisiae) (218193_s_at), score: -0.65 H1FXH1 histone family, member X (204805_s_at), score: 0.68 HBS1LHBS1-like (S. cerevisiae) (209314_s_at), score: -0.62 HDGFhepatoma-derived growth factor (high-mobility group protein 1-like) (200896_x_at), score: 0.74 HISPPD1histidine acid phosphatase domain containing 1 (203253_s_at), score: -0.82 HOXC5homeobox C5 (206739_at), score: -0.65 HSD17B6hydroxysteroid (17-beta) dehydrogenase 6 homolog (mouse) (37512_at), score: -0.8 IBTKinhibitor of Bruton agammaglobulinemia tyrosine kinase (210970_s_at), score: -0.68 IL33interleukin 33 (209821_at), score: -0.66 INSIG2insulin induced gene 2 (209566_at), score: -0.64 ITGAVintegrin, alpha V (vitronectin receptor, alpha polypeptide, antigen CD51) (202351_at), score: -0.67 JMJD1Cjumonji domain containing 1C (221763_at), score: -0.66 KAL1Kallmann syndrome 1 sequence (205206_at), score: -1 KCNJ8potassium inwardly-rectifying channel, subfamily J, member 8 (205303_at), score: -0.64 KCNK1potassium channel, subfamily K, member 1 (204679_at), score: -0.62 KIAA0562KIAA0562 (204075_s_at), score: -0.68 KIAA1462KIAA1462 (213316_at), score: 0.65 LARP4La ribonucleoprotein domain family, member 4 (214155_s_at), score: -0.84 LOC391132similar to hCG2041276 (216177_at), score: -0.67 LOC440434hypothetical protein FLJ11822 (215090_x_at), score: 0.63 LPGAT1lysophosphatidylglycerol acyltransferase 1 (202651_at), score: -0.64 LRP8low density lipoprotein receptor-related protein 8, apolipoprotein e receptor (205282_at), score: -0.65 LRRN3leucine rich repeat neuronal 3 (209841_s_at), score: -0.74 LYPD1LY6/PLAUR domain containing 1 (212909_at), score: -0.66 MAP3K6mitogen-activated protein kinase kinase kinase 6 (219278_at), score: 0.62 MATR3matrin 3 (200626_s_at), score: -0.63 MED9mediator complex subunit 9 (218372_at), score: 0.6 MFAP3microfibrillar-associated protein 3 (213123_at), score: -0.67 MFAP3Lmicrofibrillar-associated protein 3-like (205442_at), score: -0.92 MMP10matrix metallopeptidase 10 (stromelysin 2) (205680_at), score: -0.64 MSTO1misato homolog 1 (Drosophila) (218296_x_at), score: -0.63 NCALDneurocalcin delta (211685_s_at), score: -0.73 NCAPD2non-SMC condensin I complex, subunit D2 (201774_s_at), score: 0.6 NDPNorrie disease (pseudoglioma) (206022_at), score: 0.6 NEFMneurofilament, medium polypeptide (205113_at), score: -0.87 NFKBIAnuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha (201502_s_at), score: 0.62 NOX4NADPH oxidase 4 (219773_at), score: -0.77 NPIPnuclear pore complex interacting protein (204538_x_at), score: 0.61 NPTX1neuronal pentraxin I (204684_at), score: -0.65 NRXN3neurexin 3 (205795_at), score: -0.69 OBFC2Aoligonucleotide/oligosaccharide-binding fold containing 2A (219334_s_at), score: -0.64 OLFML2Bolfactomedin-like 2B (213125_at), score: 0.63 OLR1oxidized low density lipoprotein (lectin-like) receptor 1 (210004_at), score: -0.67 OPCMLopioid binding protein/cell adhesion molecule-like (214111_at), score: -0.72 OSTM1osteopetrosis associated transmembrane protein 1 (218196_at), score: -0.73 PAK1p21 protein (Cdc42/Rac)-activated kinase 1 (209615_s_at), score: 0.6 PAQR3progestin and adipoQ receptor family member III (213372_at), score: -0.74 PCDH9protocadherin 9 (219737_s_at), score: -0.91 PIONpigeon homolog (Drosophila) (222150_s_at), score: -0.67 PLAG1pleiomorphic adenoma gene 1 (205372_at), score: -0.68 PLAUplasminogen activator, urokinase (211668_s_at), score: 0.62 PNPLA2patatin-like phospholipase domain containing 2 (212705_x_at), score: 0.65 PRKCHprotein kinase C, eta (218764_at), score: 0.63 PRMT3protein arginine methyltransferase 3 (213320_at), score: -0.81 PTHLHparathyroid hormone-like hormone (211756_at), score: -0.64 PTTG1pituitary tumor-transforming 1 (203554_x_at), score: 0.61 PVRL3poliovirus receptor-related 3 (213325_at), score: -0.64 RAB1ARAB1A, member RAS oncogene family (213440_at), score: -0.83 RAB6CRAB6C, member RAS oncogene family (210406_s_at), score: -0.66 RABGGTBRab geranylgeranyltransferase, beta subunit (213704_at), score: -0.73 RANBP6RAN binding protein 6 (213019_at), score: -0.66 REREarginine-glutamic acid dipeptide (RE) repeats (200940_s_at), score: 0.75 RNF11ring finger protein 11 (208924_at), score: -0.66 RNF220ring finger protein 220 (219988_s_at), score: 0.6 RP5-1000E10.4suppressor of IKK epsilon (221705_s_at), score: -0.66 RRP15ribosomal RNA processing 15 homolog (S. cerevisiae) (219037_at), score: -0.68 SAP30LSAP30-like (219129_s_at), score: 0.72 SH3D19SH3 domain containing 19 (211620_x_at), score: 0.63 SLC25A13solute carrier family 25, member 13 (citrin) (203775_at), score: -0.63 SLC25A32solute carrier family 25, member 32 (221020_s_at), score: -0.62 SLC33A1solute carrier family 33 (acetyl-CoA transporter), member 1 (203165_s_at), score: -0.84 SLC43A3solute carrier family 43, member 3 (213113_s_at), score: 0.62 SLC4A7solute carrier family 4, sodium bicarbonate cotransporter, member 7 (209884_s_at), score: -0.9 SLMO2slowmo homolog 2 (Drosophila) (217851_s_at), score: -0.91 SMAD3SMAD family member 3 (218284_at), score: 0.77 SNNstannin (218032_at), score: 0.81 SSTR1somatostatin receptor 1 (208482_at), score: -0.78 SYNE1spectrin repeat containing, nuclear envelope 1 (209447_at), score: -0.76 SYNJ1synaptojanin 1 (212990_at), score: -0.7 TAF13TAF13 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 18kDa (205966_at), score: -0.67 TAF2TAF2 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 150kDa (209523_at), score: -0.7 TASP1taspase, threonine aspartase, 1 (219443_at), score: -0.63 TCEB1transcription elongation factor B (SIII), polypeptide 1 (15kDa, elongin C) (202823_at), score: -0.7 TFDP2transcription factor Dp-2 (E2F dimerization partner 2) (203588_s_at), score: 0.68 TGDSTDP-glucose 4,6-dehydratase (208249_s_at), score: -0.81 THSD7Athrombospondin, type I, domain containing 7A (214920_at), score: 0.65 TICAM2toll-like receptor adaptor molecule 2 (214658_at), score: -0.82 TM9SF3transmembrane 9 superfamily member 3 (217758_s_at), score: -0.65 TMED7transmembrane emp24 protein transport domain containing 7 (209404_s_at), score: -0.65 TMEFF1transmembrane protein with EGF-like and two follistatin-like domains 1 (205122_at), score: -0.63 TMEM30Atransmembrane protein 30A (217743_s_at), score: -0.7 TMF1TATA element modulatory factor 1 (213024_at), score: -0.73 TMSB15Bthymosin beta 15B (214051_at), score: 0.59 TOMM20translocase of outer mitochondrial membrane 20 homolog (yeast) (200662_s_at), score: -0.64 TPBGtrophoblast glycoprotein (203476_at), score: 0.62 TRAFD1TRAF-type zinc finger domain containing 1 (35254_at), score: 0.61 TRAPPC2trafficking protein particle complex 2 (219351_at), score: -0.66 TRIM21tripartite motif-containing 21 (204804_at), score: 0.71 TRPC4transient receptor potential cation channel, subfamily C, member 4 (220817_at), score: -0.64 TSNAXtranslin-associated factor X (203983_at), score: -0.63 UBE2Wubiquitin-conjugating enzyme E2W (putative) (218521_s_at), score: -0.63 UFM1ubiquitin-fold modifier 1 (218050_at), score: -0.76 UGDHUDP-glucose dehydrogenase (203343_at), score: -0.69 URB1URB1 ribosome biogenesis 1 homolog (S. cerevisiae) (212996_s_at), score: -0.78 VPS13Cvacuolar protein sorting 13 homolog C (S. cerevisiae) (218396_at), score: -0.65 WDR6WD repeat domain 6 (217734_s_at), score: 0.62 YDD19YDD19 protein (37079_at), score: -0.67 YTHDC2YTH domain containing 2 (213077_at), score: -0.65 ZBTB6zinc finger and BTB domain containing 6 (206098_at), score: -0.65 ZCCHC10zinc finger, CCHC domain containing 10 (221193_s_at), score: -0.63 ZDHHC7zinc finger, DHHC-type containing 7 (218606_at), score: 0.89 ZNF23zinc finger protein 23 (KOX 16) (213934_s_at), score: -0.7
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
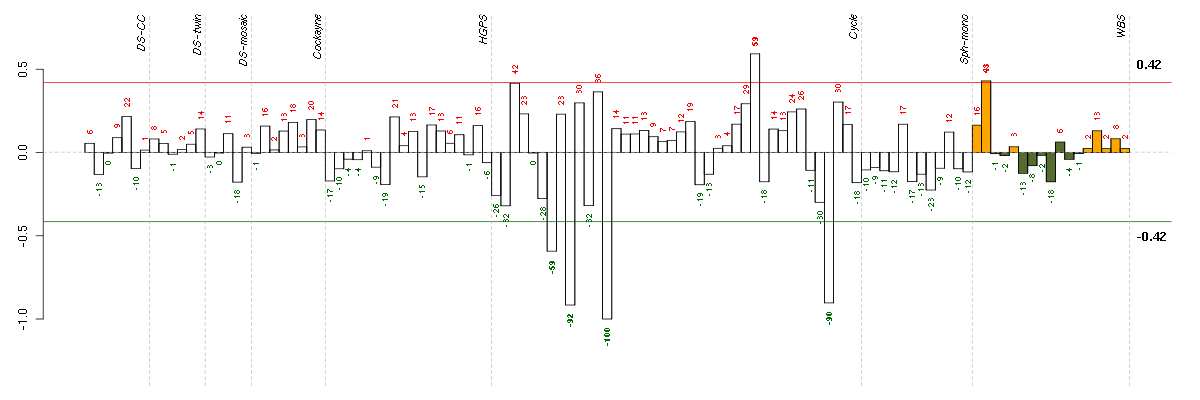
| E-TABM-263-raw-cel-1515485891.cel | 13 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485811.cel | 9 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486371.cel | 37 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485771.cel | 7 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| 10590_WBS.CEL | 2 | 8 | WBS | hgu133plus2 | WBS | WBS 1 |
| E-TABM-263-raw-cel-1515486211.cel | 29 | 6 | Cycle | hgu133a2 | none | Cycle 1 |