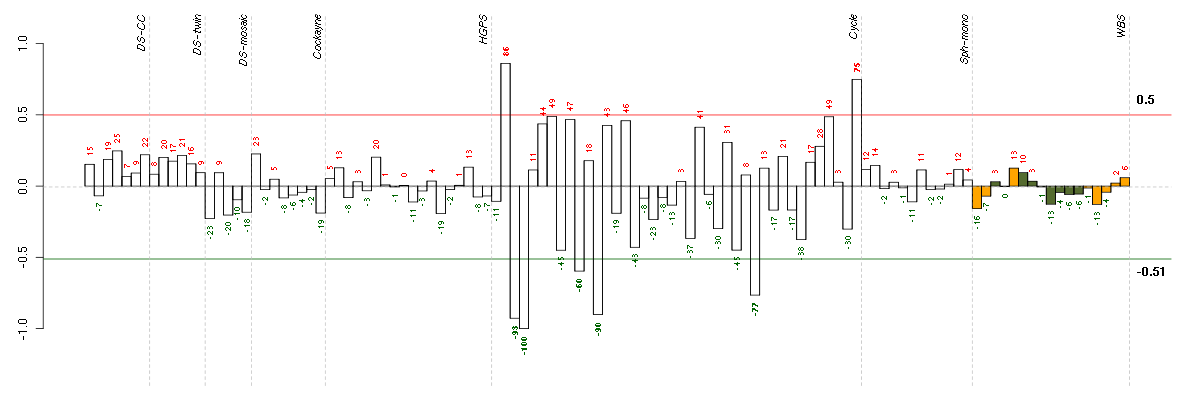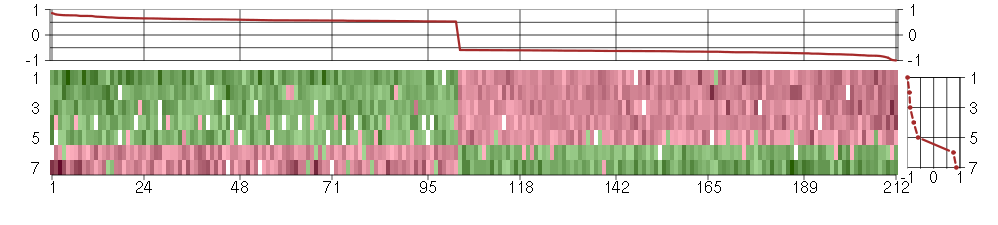

Under-expression is coded with green,
over-expression with red color.



ABCA7ATP-binding cassette, sub-family A (ABC1), member 7 (219577_s_at), score: 0.79 ABCC9ATP-binding cassette, sub-family C (CFTR/MRP), member 9 (208561_at), score: -0.63 ABCG4ATP-binding cassette, sub-family G (WHITE), member 4 (207593_at), score: 0.55 ABLIM1actin binding LIM protein 1 (200965_s_at), score: -0.65 ADAMTS7ADAM metallopeptidase with thrombospondin type 1 motif, 7 (220705_s_at), score: 0.57 ADORA1adenosine A1 receptor (216220_s_at), score: 0.63 AGTangiotensinogen (serpin peptidase inhibitor, clade A, member 8) (202834_at), score: 0.57 ALDH1L1aldehyde dehydrogenase 1 family, member L1 (215798_at), score: 0.71 ALDH3A1aldehyde dehydrogenase 3 family, memberA1 (205623_at), score: 0.56 ANKFY1ankyrin repeat and FYVE domain containing 1 (219868_s_at), score: -0.63 APPamyloid beta (A4) precursor protein (214953_s_at), score: 0.63 ARHGAP22Rho GTPase activating protein 22 (206298_at), score: -0.6 ARHGDIARho GDP dissociation inhibitor (GDI) alpha (213606_s_at), score: -0.62 ASPHD1aspartate beta-hydroxylase domain containing 1 (214993_at), score: 0.57 ATF5activating transcription factor 5 (204999_s_at), score: -0.62 ATMataxia telangiectasia mutated (210858_x_at), score: 0.57 ATP6V0A1ATPase, H+ transporting, lysosomal V0 subunit a1 (205095_s_at), score: -0.69 B3GAT3beta-1,3-glucuronyltransferase 3 (glucuronosyltransferase I) (203452_at), score: -0.6 BAP1BRCA1 associated protein-1 (ubiquitin carboxy-terminal hydrolase) (201419_at), score: -0.63 BAT2HLA-B associated transcript 2 (212081_x_at), score: -0.7 BAT2LHLA-B associated transcript 2-like (212068_s_at), score: -0.6 BCAMbasal cell adhesion molecule (Lutheran blood group) (40093_at), score: 0.59 BCL2L1BCL2-like 1 (215037_s_at), score: -0.68 BTBD2BTB (POZ) domain containing 2 (207722_s_at), score: -0.63 BTNL3butyrophilin-like 3 (217207_s_at), score: 0.54 C13orf34chromosome 13 open reading frame 34 (219544_at), score: -0.62 C19orf61chromosome 19 open reading frame 61 (221335_x_at), score: -0.62 C20orf111chromosome 20 open reading frame 111 (209020_at), score: 0.53 C20orf117chromosome 20 open reading frame 117 (207711_at), score: -0.63 C2orf37chromosome 2 open reading frame 37 (220172_at), score: -0.79 CALB2calbindin 2 (205428_s_at), score: 0.6 CAPN5calpain 5 (205166_at), score: -0.59 CCDC40coiled-coil domain containing 40 (220592_at), score: 0.62 CD79BCD79b molecule, immunoglobulin-associated beta (205297_s_at), score: 0.8 CDC2L6cell division cycle 2-like 6 (CDK8-like) (212899_at), score: -0.63 CDC42BPBCDC42 binding protein kinase beta (DMPK-like) (217849_s_at), score: -0.7 CDC42EP4CDC42 effector protein (Rho GTPase binding) 4 (214721_x_at), score: -0.6 CHRNB2cholinergic receptor, nicotinic, beta 2 (neuronal) (206635_at), score: 0.64 CITcitron (rho-interacting, serine/threonine kinase 21) (212801_at), score: -0.62 CMAHcytidine monophosphate-N-acetylneuraminic acid hydroxylase (CMP-N-acetylneuraminate monooxygenase) pseudogene (205518_s_at), score: -0.67 CNNM1cyclin M1 (220166_at), score: 0.55 CNNM3cyclin M3 (220739_s_at), score: -0.6 CNOT3CCR4-NOT transcription complex, subunit 3 (203239_s_at), score: -0.74 COL11A2collagen, type XI, alpha 2 (216993_s_at), score: 0.55 COX6A2cytochrome c oxidase subunit VIa polypeptide 2 (206353_at), score: 0.58 CXorf21chromosome X open reading frame 21 (220252_x_at), score: 0.57 CYP3A4cytochrome P450, family 3, subfamily A, polypeptide 4 (205998_x_at), score: 0.69 DLSTPdihydrolipoamide S-succinyltransferase pseudogene (E2 component of 2-oxo-glutarate complex) (215210_s_at), score: 0.54 DMBT1deleted in malignant brain tumors 1 (208250_s_at), score: 0.77 DMPKdystrophia myotonica-protein kinase (37996_s_at), score: -0.69 DUSP9dual specificity phosphatase 9 (205777_at), score: 0.61 EDEM1ER degradation enhancer, mannosidase alpha-like 1 (203279_at), score: 0.54 ELK1ELK1, member of ETS oncogene family (210376_x_at), score: -0.6 EMCNendomucin (219436_s_at), score: -0.98 EPS8L1EPS8-like 1 (218778_x_at), score: 0.53 ETV7ets variant 7 (221680_s_at), score: 0.57 EVI2Becotropic viral integration site 2B (211742_s_at), score: -0.65 EXOC2exocyst complex component 2 (219349_s_at), score: -0.72 FAM105Afamily with sequence similarity 105, member A (219694_at), score: -0.73 FBP1fructose-1,6-bisphosphatase 1 (209696_at), score: 0.77 FBXL14F-box and leucine-rich repeat protein 14 (213145_at), score: -0.64 FBXO2F-box protein 2 (219305_x_at), score: 0.53 FER1L4fer-1-like 4 (C. elegans) (222245_s_at), score: 0.58 FKBP10FK506 binding protein 10, 65 kDa (219249_s_at), score: 0.75 FKRPfukutin related protein (219853_at), score: -0.7 FLRT3fibronectin leucine rich transmembrane protein 3 (219250_s_at), score: -0.68 FOXC2forkhead box C2 (MFH-1, mesenchyme forkhead 1) (214520_at), score: -0.63 FUT2fucosyltransferase 2 (secretor status included) (210608_s_at), score: 0.61 GABARAPL1GABA(A) receptor-associated protein like 1 (208868_s_at), score: 0.6 GABARAPL3GABA(A) receptors associated protein like 3 (pseudogene) (211458_s_at), score: 0.63 GBA3glucosidase, beta, acid 3 (cytosolic) (219954_s_at), score: 0.78 GDF9growth differentiation factor 9 (221314_at), score: 0.66 GFOD1glucose-fructose oxidoreductase domain containing 1 (219821_s_at), score: 0.64 GGA3golgi associated, gamma adaptin ear containing, ARF binding protein 3 (209411_s_at), score: -0.61 GMPRguanosine monophosphate reductase (204187_at), score: -0.6 GNL3Lguanine nucleotide binding protein-like 3 (nucleolar)-like (205010_at), score: 0.54 GPR56G protein-coupled receptor 56 (212070_at), score: -0.7 H1FXH1 histone family, member X (204805_s_at), score: -0.62 HAB1B1 for mucin (215778_x_at), score: 0.74 HAMPhepcidin antimicrobial peptide (220491_at), score: 0.65 HDAC5histone deacetylase 5 (202455_at), score: -0.59 HGFhepatocyte growth factor (hepapoietin A; scatter factor) (209960_at), score: -0.64 HIST1H1Chistone cluster 1, H1c (209398_at), score: -0.59 HIST1H4Ghistone cluster 1, H4g (208551_at), score: 0.86 HMGA1high mobility group AT-hook 1 (210457_x_at), score: -0.73 HSD17B6hydroxysteroid (17-beta) dehydrogenase 6 homolog (mouse) (37512_at), score: 0.55 HSPA4Lheat shock 70kDa protein 4-like (205543_at), score: -0.71 HUNKhormonally up-regulated Neu-associated kinase (219535_at), score: 0.54 IDUAiduronidase, alpha-L- (205059_s_at), score: -0.63 INAinternexin neuronal intermediate filament protein, alpha (204465_s_at), score: -0.68 IRF2interferon regulatory factor 2 (203275_at), score: -0.72 ITGALintegrin, alpha L (antigen CD11A (p180), lymphocyte function-associated antigen 1; alpha polypeptide) (213475_s_at), score: 0.57 JAK2Janus kinase 2 (a protein tyrosine kinase) (205842_s_at), score: -0.63 JUPjunction plakoglobin (201015_s_at), score: -0.75 KCTD15potassium channel tetramerisation domain containing 15 (218553_s_at), score: 0.6 KIAA1305KIAA1305 (220911_s_at), score: -0.73 LEPREL2leprecan-like 2 (204854_at), score: -0.63 LILRB3leukocyte immunoglobulin-like receptor, subfamily B (with TM and ITIM domains), member 3 (211133_x_at), score: 0.55 LLGL2lethal giant larvae homolog 2 (Drosophila) (203713_s_at), score: 0.55 LOC149501similar to keratin 8 (216821_at), score: 0.62 LOC391132similar to hCG2041276 (216177_at), score: 0.56 LOC440434hypothetical protein FLJ11822 (215090_x_at), score: -0.61 LOC730227hypothetical protein LOC730227 (215756_at), score: 0.53 LOC80054hypothetical LOC80054 (220465_at), score: 0.53 LPHN1latrophilin 1 (203488_at), score: 0.57 LRDDleucine-rich repeats and death domain containing (221640_s_at), score: -0.62 LRP2low density lipoprotein-related protein 2 (205710_at), score: -0.65 LRP8low density lipoprotein receptor-related protein 8, apolipoprotein e receptor (205282_at), score: 0.53 MALmal, T-cell differentiation protein (204777_s_at), score: 0.53 MAP3K6mitogen-activated protein kinase kinase kinase 6 (219278_at), score: -0.68 MDM2Mdm2 p53 binding protein homolog (mouse) (205386_s_at), score: -0.77 MEOX1mesenchyme homeobox 1 (205619_s_at), score: 0.66 MFNGMFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase (204153_s_at), score: 0.61 MMP17matrix metallopeptidase 17 (membrane-inserted) (206234_s_at), score: 0.53 MOBPmyelin-associated oligodendrocyte basic protein (210193_at), score: 0.77 MOCS1molybdenum cofactor synthesis 1 (211673_s_at), score: 0.67 MOGmyelin oligodendrocyte glycoprotein (214650_x_at), score: 0.61 MPP1membrane protein, palmitoylated 1, 55kDa (202974_at), score: -0.66 MRPS6mitochondrial ribosomal protein S6 (213167_s_at), score: -0.81 MTMR11myotubularin related protein 11 (205076_s_at), score: -0.75 MUC13mucin 13, cell surface associated (218687_s_at), score: 0.59 MUC5ACmucin 5AC, oligomeric mucus/gel-forming (217182_at), score: 0.61 MUL1mitochondrial E3 ubiquitin ligase 1 (218246_at), score: -0.59 MYH15myosin, heavy chain 15 (215331_at), score: 0.57 MYL4myosin, light chain 4, alkali; atrial, embryonic (210088_x_at), score: 0.65 NACAP1nascent-polypeptide-associated complex alpha polypeptide pseudogene 1 (211445_x_at), score: 0.58 NCOA2nuclear receptor coactivator 2 (205732_s_at), score: -0.61 NDPNorrie disease (pseudoglioma) (206022_at), score: -0.81 NEFMneurofilament, medium polypeptide (205113_at), score: 0.62 NEO1neogenin homolog 1 (chicken) (204321_at), score: -0.59 NF1neurofibromin 1 (211094_s_at), score: -0.89 NFATC4nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 4 (205897_at), score: -0.6 NFYCnuclear transcription factor Y, gamma (202215_s_at), score: -0.68 NOVA2neuro-oncological ventral antigen 2 (206477_s_at), score: 0.59 NUPL2nucleoporin like 2 (204003_s_at), score: 0.61 NXPH3neurexophilin 3 (221991_at), score: 0.65 OAS32'-5'-oligoadenylate synthetase 3, 100kDa (218400_at), score: -0.69 OLFML2Bolfactomedin-like 2B (213125_at), score: -1 OR7E47Polfactory receptor, family 7, subfamily E, member 47 pseudogene (216698_x_at), score: 0.56 PAK1p21 protein (Cdc42/Rac)-activated kinase 1 (209615_s_at), score: -0.82 PARD6Apar-6 partitioning defective 6 homolog alpha (C. elegans) (205245_at), score: 0.57 PARVBparvin, beta (204629_at), score: -0.65 PCDHGA11protocadherin gamma subfamily A, 11 (211876_x_at), score: -0.61 PCYT2phosphate cytidylyltransferase 2, ethanolamine (209577_at), score: -0.61 PDE5Aphosphodiesterase 5A, cGMP-specific (206757_at), score: -0.85 PDPRpyruvate dehydrogenase phosphatase regulatory subunit (220236_at), score: -0.63 PHF7PHD finger protein 7 (215622_x_at), score: 0.58 PIGOphosphatidylinositol glycan anchor biosynthesis, class O (209998_at), score: -0.62 PMLpromyelocytic leukemia (206503_x_at), score: -0.66 PNMAL1PNMA-like 1 (218824_at), score: -0.67 POM121L2POM121 membrane glycoprotein-like 2 (rat) (216582_at), score: 0.57 PPP1R13Bprotein phosphatase 1, regulatory (inhibitor) subunit 13B (216347_s_at), score: 0.62 PRLHprolactin releasing hormone (221443_x_at), score: 0.72 PRMT3protein arginine methyltransferase 3 (213320_at), score: 0.55 PTPN20Bprotein tyrosine phosphatase, non-receptor type 20B (215172_at), score: -0.63 PTPN6protein tyrosine phosphatase, non-receptor type 6 (206687_s_at), score: 0.75 PXNpaxillin (211823_s_at), score: -0.81 RASGRP3RAS guanyl releasing protein 3 (calcium and DAG-regulated) (205801_s_at), score: -0.68 RBM38RNA binding motif protein 38 (212430_at), score: 0.57 REG3Aregenerating islet-derived 3 alpha (205815_at), score: 0.71 RNF220ring finger protein 220 (219988_s_at), score: -0.63 RP4-691N24.1ninein-like (207705_s_at), score: -0.71 RPL18AP6ribosomal protein L18a pseudogene 6 (216383_at), score: 0.62 RPRD2regulation of nuclear pre-mRNA domain containing 2 (212553_at), score: -0.69 RRP15ribosomal RNA processing 15 homolog (S. cerevisiae) (219037_at), score: 0.56 RSAD1radical S-adenosyl methionine domain containing 1 (218307_at), score: -0.65 RXRBretinoid X receptor, beta (215099_s_at), score: -0.61 SAA4serum amyloid A4, constitutive (207096_at), score: 0.65 SAV1salvador homolog 1 (Drosophila) (218276_s_at), score: -0.67 SCAMP4secretory carrier membrane protein 4 (213244_at), score: -0.76 SDPRserum deprivation response (phosphatidylserine binding protein) (218711_s_at), score: -0.77 SEC61A2Sec61 alpha 2 subunit (S. cerevisiae) (219499_at), score: -0.71 SEMA3Fsema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3F (35666_at), score: 0.57 SENP7SUMO1/sentrin specific peptidase 7 (220735_s_at), score: -0.66 SF1splicing factor 1 (208313_s_at), score: -0.61 SH3GLB2SH3-domain GRB2-like endophilin B2 (218813_s_at), score: -0.59 SIRT6sirtuin (silent mating type information regulation 2 homolog) 6 (S. cerevisiae) (219613_s_at), score: -0.65 SLC12A6solute carrier family 12 (potassium/chloride transporters), member 6 (220740_s_at), score: -0.59 SLC26A10solute carrier family 26, member 10 (214951_at), score: 0.65 SLC5A5solute carrier family 5 (sodium iodide symporter), member 5 (211123_at), score: 0.68 SLC6A11solute carrier family 6 (neurotransmitter transporter, GABA), member 11 (207048_at), score: 0.66 SMG7Smg-7 homolog, nonsense mediated mRNA decay factor (C. elegans) (201794_s_at), score: -0.59 SNAP25synaptosomal-associated protein, 25kDa (202508_s_at), score: -0.65 SOLHsmall optic lobes homolog (Drosophila) (204275_at), score: -0.64 SPAG8sperm associated antigen 8 (206816_s_at), score: 0.67 SPATA1spermatogenesis associated 1 (221057_at), score: 0.64 SPG7spastic paraplegia 7 (pure and complicated autosomal recessive) (202104_s_at), score: -0.61 SSX3synovial sarcoma, X breakpoint 3 (211731_x_at), score: 0.6 STIM1stromal interaction molecule 1 (202764_at), score: -0.8 SULT1B1sulfotransferase family, cytosolic, 1B, member 1 (207601_at), score: 0.69 TGFBR2transforming growth factor, beta receptor II (70/80kDa) (207334_s_at), score: -0.75 TGIF2TGFB-induced factor homeobox 2 (216262_s_at), score: 0.57 TGM4transglutaminase 4 (prostate) (217566_s_at), score: 0.75 TIAF1TGFB1-induced anti-apoptotic factor 1 (202039_at), score: -0.63 TLE3transducin-like enhancer of split 3 (E(sp1) homolog, Drosophila) (206472_s_at), score: -0.61 TMPRSS6transmembrane protease, serine 6 (214955_at), score: 0.55 TMSB4Ythymosin beta 4, Y-linked (206769_at), score: 0.57 TNK1tyrosine kinase, non-receptor, 1 (217149_x_at), score: 0.58 TOM1L2target of myb1-like 2 (chicken) (214840_at), score: -0.64 TP53tumor protein p53 (201746_at), score: -0.6 TRMT2ATRM2 tRNA methyltransferase 2 homolog A (S. cerevisiae) (91617_at), score: 0.54 TTC33tetratricopeptide repeat domain 33 (219421_at), score: -0.6 TUBA4Btubulin, alpha 4b (pseudogene) (207490_at), score: 0.56 TULP2tubby like protein 2 (206733_at), score: 0.63 UBTD1ubiquitin domain containing 1 (219172_at), score: -0.76 ULK1unc-51-like kinase 1 (C. elegans) (209333_at), score: -0.6 UTF1undifferentiated embryonic cell transcription factor 1 (208275_x_at), score: 0.55 VEGFBvascular endothelial growth factor B (203683_s_at), score: -0.78 WDR42AWD repeat domain 42A (202249_s_at), score: -0.66 WDR6WD repeat domain 6 (217734_s_at), score: -0.6 YAF2YY1 associated factor 2 (206238_s_at), score: 0.53 ZNF783zinc finger family member 783 (221876_at), score: 0.55
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-TABM-263-raw-cel-1515485711.cel | 4 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485691.cel | 3 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485871.cel | 12 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486211.cel | 29 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485831.cel | 10 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486431.cel | 40 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485671.cel | 2 | 6 | Cycle | hgu133a2 | none | Cycle 1 |