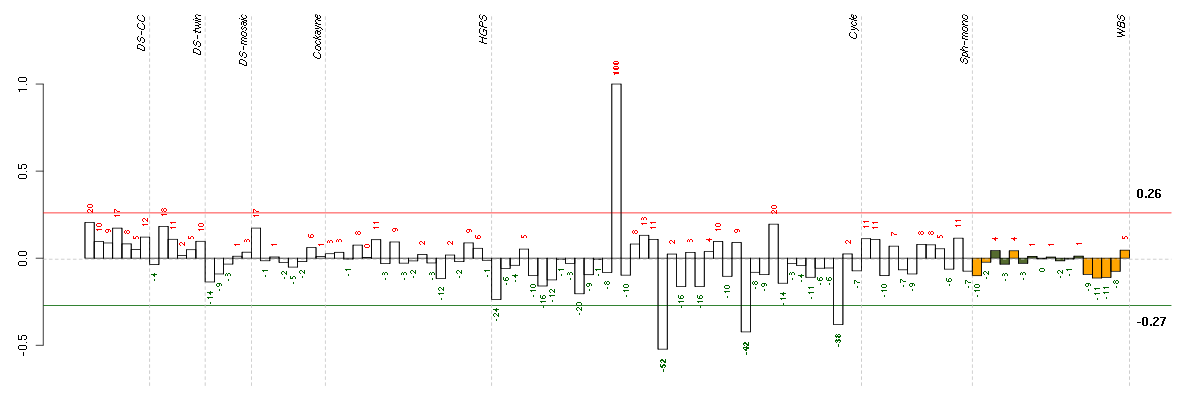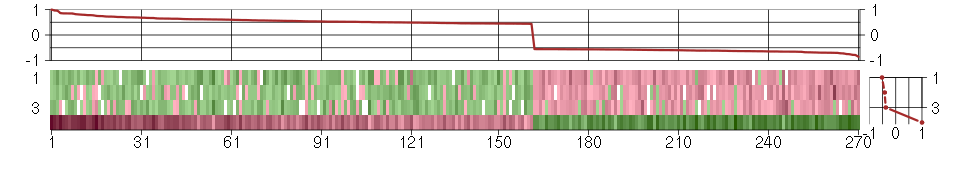

Under-expression is coded with green,
over-expression with red color.


plasma membrane
The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins.
membrane
Double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
integral to membrane
Penetrating at least one phospholipid bilayer of a membrane. May also refer to the state of being buried in the bilayer with no exposure outside the bilayer. When used to describe a protein, indicates that all or part of the peptide sequence is embedded in the membrane.
integral to plasma membrane
Penetrating at least one phospholipid bilayer of a plasma membrane. May also refer to the state of being buried in the bilayer with no exposure outside the bilayer.
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
cell
The basic structural and functional unit of all organisms. Includes the plasma membrane and any external encapsulating structures such as the cell wall and cell envelope.
intrinsic to membrane
Located in a membrane such that some covalently attached portion of the gene product, for example part of a peptide sequence or some other covalently attached moiety such as a GPI anchor, spans or is embedded in one or both leaflets of the membrane.
intrinsic to plasma membrane
Located in the plasma membrane such that some covalently attached portion of the gene product, for example part of a peptide sequence or some other covalently attached moiety such as a GPI anchor, spans or is embedded in one or both leaflets of the membrane.
membrane part
Any constituent part of a membrane, a double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
plasma membrane part
Any constituent part of the plasma membrane, the membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins.
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
all
This term is the most general term possible
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
membrane part
Any constituent part of a membrane, a double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
plasma membrane part
Any constituent part of the plasma membrane, the membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins.
intrinsic to plasma membrane
Located in the plasma membrane such that some covalently attached portion of the gene product, for example part of a peptide sequence or some other covalently attached moiety such as a GPI anchor, spans or is embedded in one or both leaflets of the membrane.
integral to plasma membrane
Penetrating at least one phospholipid bilayer of a plasma membrane. May also refer to the state of being buried in the bilayer with no exposure outside the bilayer.

molecular_function
Elemental activities, such as catalysis or binding, describing the actions of a gene product at the molecular level. A given gene product may exhibit one or more molecular functions.
binding
The selective, often stoichiometric, interaction of a molecule with one or more specific sites on another molecule.
metal ion binding
Interacting selectively with any metal ion.
alkali metal ion binding
Interacting selectively with any alkali metal ion; alkali metals are those elements in group Ia of the periodic table, with the exception of hydrogen.
ion binding
Interacting selectively with ions, charged atoms or groups of atoms.
cation binding
Interacting selectively with cations, charged atoms or groups of atoms with a net positive charge.
all
This term is the most general term possible
alkali metal ion binding
Interacting selectively with any alkali metal ion; alkali metals are those elements in group Ia of the periodic table, with the exception of hydrogen.
ABT1activator of basal transcription 1 (218405_at), score: -0.58 ABTB2ankyrin repeat and BTB (POZ) domain containing 2 (213497_at), score: -0.64 ACANaggrecan (205679_x_at), score: -0.56 ACSL6acyl-CoA synthetase long-chain family member 6 (211207_s_at), score: 0.64 ADAMDEC1ADAM-like, decysin 1 (206134_at), score: 0.82 ADAMTS6ADAM metallopeptidase with thrombospondin type 1 motif, 6 (220866_at), score: 0.52 ADAMTS9ADAM metallopeptidase with thrombospondin type 1 motif, 9 (220287_at), score: 0.67 ADCK2aarF domain containing kinase 2 (221893_s_at), score: 0.59 AGAP2ArfGAP with GTPase domain, ankyrin repeat and PH domain 2 (215080_s_at), score: 0.52 AKR1D1aldo-keto reductase family 1, member D1 (delta 4-3-ketosteroid-5-beta-reductase) (207102_at), score: 0.52 AMBRA1autophagy/beclin-1 regulator 1 (52731_at), score: -0.61 ANKS1Aankyrin repeat and sterile alpha motif domain containing 1A (212747_at), score: -0.66 ANO1anoctamin 1, calcium activated chloride channel (218804_at), score: 0.45 ARHGEF18rho/rac guanine nucleotide exchange factor (GEF) 18 (213039_at), score: -0.56 ASTN2astrotactin 2 (209693_at), score: -0.78 ATAD5ATPase family, AAA domain containing 5 (220223_at), score: 0.47 ATHL1ATH1, acid trehalase-like 1 (yeast) (219359_at), score: 0.54 ATP1B4ATPase, (Na+)/K+ transporting, beta 4 polypeptide (220556_at), score: 0.45 AVILadvillin (205539_at), score: -0.6 BBS10Bardet-Biedl syndrome 10 (219487_at), score: -0.59 BCL2B-cell CLL/lymphoma 2 (203685_at), score: 0.55 BCS1LBCS1-like (yeast) (207618_s_at), score: -0.55 BICC1bicaudal C homolog 1 (Drosophila) (220580_at), score: 0.51 BTN3A1butyrophilin, subfamily 3, member A1 (209770_at), score: -0.56 C12orf5chromosome 12 open reading frame 5 (219099_at), score: -0.56 C17orf39chromosome 17 open reading frame 39 (220058_at), score: -0.7 C1orf115chromosome 1 open reading frame 115 (218546_at), score: 0.57 C1orf66chromosome 1 open reading frame 66 (218914_at), score: -0.57 C3AR1complement component 3a receptor 1 (209906_at), score: -0.68 C3orf51chromosome 3 open reading frame 51 (208247_at), score: 0.52 C4orf16chromosome 4 open reading frame 16 (219023_at), score: -0.57 C4orf29chromosome 4 open reading frame 29 (219980_at), score: 0.71 C5orf44chromosome 5 open reading frame 44 (218674_at), score: -0.57 CAND2cullin-associated and neddylation-dissociated 2 (putative) (213547_at), score: -0.63 CD200CD200 molecule (209583_s_at), score: 0.64 CD226CD226 molecule (207315_at), score: 0.49 CD28CD28 molecule (206545_at), score: 0.65 CD320CD320 molecule (218529_at), score: -0.63 CD40CD40 molecule, TNF receptor superfamily member 5 (215346_at), score: -0.61 CDH18cadherin 18, type 2 (206280_at), score: -0.56 CDH19cadherin 19, type 2 (206898_at), score: 0.75 CDK10cyclin-dependent kinase 10 (203468_at), score: -0.56 CEP70centrosomal protein 70kDa (219036_at), score: -0.63 CHKAcholine kinase alpha (204233_s_at), score: -0.59 CHN2chimerin (chimaerin) 2 (207486_x_at), score: 0.78 CHRNA5cholinergic receptor, nicotinic, alpha 5 (206533_at), score: 0.61 CIRCBF1 interacting corepressor (209571_at), score: -0.56 CLCN2chloride channel 2 (213499_at), score: -0.61 CLEC1AC-type lectin domain family 1, member A (219761_at), score: 0.56 CLEC2DC-type lectin domain family 2, member D (220132_s_at), score: 0.53 CLTCL1clathrin, heavy chain-like 1 (205944_s_at), score: -0.62 COL10A1collagen, type X, alpha 1 (217428_s_at), score: 0.45 COPS7BCOP9 constitutive photomorphogenic homolog subunit 7B (Arabidopsis) (219997_s_at), score: -0.65 COQ3coenzyme Q3 homolog, methyltransferase (S. cerevisiae) (221227_x_at), score: -0.63 COQ7coenzyme Q7 homolog, ubiquinone (yeast) (209745_at), score: 0.56 CORO2Bcoronin, actin binding protein, 2B (209789_at), score: 0.62 CPA4carboxypeptidase A4 (205832_at), score: -0.57 CPT2carnitine palmitoyltransferase 2 (204264_at), score: -0.64 CTAGE4CTAGE family, member 4 (215549_x_at), score: -0.65 CTNNBIP1catenin, beta interacting protein 1 (203081_at), score: 0.49 CTSScathepsin S (202901_x_at), score: 0.95 CXCL11chemokine (C-X-C motif) ligand 11 (210163_at), score: 0.56 CYCSP52cytochrome c, somatic pseudogene 52 (217206_at), score: 0.8 CYP26B1cytochrome P450, family 26, subfamily B, polypeptide 1 (219825_at), score: 0.61 CYSLTR1cysteinyl leukotriene receptor 1 (216288_at), score: 0.5 DCUN1D2DCN1, defective in cullin neddylation 1, domain containing 2 (S. cerevisiae) (219116_s_at), score: -0.68 DIRAS3DIRAS family, GTP-binding RAS-like 3 (215506_s_at), score: 0.44 DNAL4dynein, axonemal, light chain 4 (204008_at), score: -0.56 DSCR6Down syndrome critical region gene 6 (207267_s_at), score: 0.74 DSG2desmoglein 2 (217901_at), score: 0.52 DSTYKdual serine/threonine and tyrosine protein kinase (214663_at), score: -0.62 DTNAdystrobrevin, alpha (205741_s_at), score: 0.61 DTNBdystrobrevin, beta (215295_at), score: 0.77 EDA2Rectodysplasin A2 receptor (221399_at), score: 0.47 ELLelongation factor RNA polymerase II (204096_s_at), score: -0.57 EML2echinoderm microtubule associated protein like 2 (204398_s_at), score: -0.63 ENPEPglutamyl aminopeptidase (aminopeptidase A) (204844_at), score: 0.58 EPOerythropoietin (217254_s_at), score: -0.7 ERBB3v-erb-b2 erythroblastic leukemia viral oncogene homolog 3 (avian) (202454_s_at), score: 0.5 ERBB4v-erb-a erythroblastic leukemia viral oncogene homolog 4 (avian) (214053_at), score: 0.72 ERN1endoplasmic reticulum to nucleus signaling 1 (207061_at), score: 0.62 EVI2Becotropic viral integration site 2B (211742_s_at), score: -0.59 EXOSC5exosome component 5 (218481_at), score: -0.69 FAM117Afamily with sequence similarity 117, member A (221249_s_at), score: -0.56 FAM169Afamily with sequence similarity 169, member A (213954_at), score: 0.61 FAM59Afamily with sequence similarity 59, member A (219377_at), score: 0.51 FAM63Bfamily with sequence similarity 63, member B (214691_x_at), score: 0.46 FBXO22F-box protein 22 (219638_at), score: 0.59 FCGR2AFc fragment of IgG, low affinity IIa, receptor (CD32) (203561_at), score: 0.57 FKRPfukutin related protein (219853_at), score: 0.54 FMO3flavin containing monooxygenase 3 (40665_at), score: 0.45 FOXE1forkhead box E1 (thyroid transcription factor 2) (206912_at), score: -0.63 FOXK2forkhead box K2 (203064_s_at), score: 0.54 FRMD4BFERM domain containing 4B (213056_at), score: 0.85 GCAgrancalcin, EF-hand calcium binding protein (203765_at), score: 0.45 GIT1G protein-coupled receptor kinase interacting ArfGAP 1 (218030_at), score: 0.45 GNA12guanine nucleotide binding protein (G protein) alpha 12 (221737_at), score: -0.57 GPR4G protein-coupled receptor 4 (206236_at), score: 0.47 GPR63G protein-coupled receptor 63 (220993_s_at), score: 0.58 GPR65G protein-coupled receptor 65 (214467_at), score: 0.45 GPRASP1G protein-coupled receptor associated sorting protein 1 (204793_at), score: -0.85 GPX3glutathione peroxidase 3 (plasma) (214091_s_at), score: 0.63 GSTCDglutathione S-transferase, C-terminal domain containing (220063_at), score: 0.96 GTF2H3general transcription factor IIH, polypeptide 3, 34kDa (222104_x_at), score: -0.56 GYG2glycogenin 2 (215695_s_at), score: -0.56 GYPCglycophorin C (Gerbich blood group) (202947_s_at), score: -0.56 HAVCR1hepatitis A virus cellular receptor 1 (207052_at), score: 0.56 HCG8HLA complex group 8 (215985_at), score: 1 HINFPhistone H4 transcription factor (206495_s_at), score: -0.68 HLA-DRB1major histocompatibility complex, class II, DR beta 1 (209312_x_at), score: 0.62 HOXB6homeobox B6 (205366_s_at), score: 0.6 HPCAL4hippocalcin like 4 (219671_at), score: 0.85 HRASLSHRAS-like suppressor (219983_at), score: 0.67 HSPBAP1HSPB (heat shock 27kDa) associated protein 1 (219284_at), score: -0.57 HTN1histatin 1 (206639_x_at), score: 0.5 HYMAIhydatidiform mole associated and imprinted (non-protein coding) (215513_at), score: 0.6 ICA1islet cell autoantigen 1, 69kDa (211740_at), score: 0.72 IFNGR1interferon gamma receptor 1 (211676_s_at), score: -0.69 IFT140intraflagellar transport 140 homolog (Chlamydomonas) (204792_s_at), score: -0.61 IGHG1immunoglobulin heavy constant gamma 1 (G1m marker) (217039_x_at), score: -0.61 IKZF4IKAROS family zinc finger 4 (Eos) (208472_at), score: -0.65 IRF6interferon regulatory factor 6 (202597_at), score: 0.62 ISYNA1inositol-3-phosphate synthase 1 (222240_s_at), score: -0.59 JMJD1Bjumonji domain containing 1B (210878_s_at), score: -0.65 KBTBD4kelch repeat and BTB (POZ) domain containing 4 (218569_s_at), score: -0.69 KCND2potassium voltage-gated channel, Shal-related subfamily, member 2 (207103_at), score: 0.52 KCNIP1Kv channel interacting protein 1 (221307_at), score: 0.78 KCNJ14potassium inwardly-rectifying channel, subfamily J, member 14 (220776_at), score: 0.59 KCNJ16potassium inwardly-rectifying channel, subfamily J, member 16 (219564_at), score: 0.69 KIAA0415KIAA0415 (209912_s_at), score: -0.59 KIRRELkin of IRRE like (Drosophila) (220825_s_at), score: -0.58 KLF12Kruppel-like factor 12 (208467_at), score: 0.49 KLHL3kelch-like 3 (Drosophila) (221221_s_at), score: 0.53 KRT2keratin 2 (207908_at), score: 0.48 KRT24keratin 24 (220267_at), score: 0.48 KRT33Akeratin 33A (208483_x_at), score: 0.55 LAIR2leukocyte-associated immunoglobulin-like receptor 2 (207509_s_at), score: 0.5 LHX3LIM homeobox 3 (221670_s_at), score: 0.55 LILRP2leukocyte immunoglobulin-like receptor pseudogene 2 (214561_at), score: 0.52 LOC100130703similar to hCG2042168 (207596_at), score: 0.57 LOC100132247similar to Uncharacterized protein KIAA0220 (215002_at), score: 0.86 LOC100134713hypothetical protein LOC100134713 (222090_at), score: -0.61 LOC257152hypothetical protein LOC257152 (215302_at), score: 0.65 LOC390940similar to R28379_1 (213556_at), score: -0.61 LOC729806similar to hCG1725380 (217544_at), score: 0.64 LOC93432maltase-glucoamylase-like pseudogene (216666_at), score: 0.64 LRP3low density lipoprotein receptor-related protein 3 (204381_at), score: -0.62 MANBAmannosidase, beta A, lysosomal (203778_at), score: -0.65 MAP1Smicrotubule-associated protein 1S (218522_s_at), score: 0.46 MAP3K9mitogen-activated protein kinase kinase kinase 9 (213927_at), score: 0.85 MAST2microtubule associated serine/threonine kinase 2 (211593_s_at), score: 0.46 MBmyoglobin (204179_at), score: 0.5 MCM3APASMCM3AP antisense RNA (non-protein coding) (220459_at), score: 0.52 MFSD6major facilitator superfamily domain containing 6 (219858_s_at), score: 0.62 MGAT4Cmannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme C (putative) (207447_s_at), score: 0.53 MGC5590hypothetical protein MGC5590 (220931_at), score: 0.58 MMP16matrix metallopeptidase 16 (membrane-inserted) (207012_at), score: 0.52 MTMR3myotubularin related protein 3 (202197_at), score: 0.57 MTUS1mitochondrial tumor suppressor 1 (212096_s_at), score: 0.47 MUSKmuscle, skeletal, receptor tyrosine kinase (207633_s_at), score: 0.45 MYBPC1myosin binding protein C, slow type (214087_s_at), score: 0.62 MYO1Amyosin IA (211916_s_at), score: 0.45 MYO5Cmyosin VC (218966_at), score: 0.81 N4BP3Nedd4 binding protein 3 (214775_at), score: 0.59 NAGPAN-acetylglucosamine-1-phosphodiester alpha-N-acetylglucosaminidase (205090_s_at), score: -0.6 NAIPNLR family, apoptosis inhibitory protein (204860_s_at), score: 0.53 NCOA1nuclear receptor coactivator 1 (209106_at), score: -0.56 NLGN4Xneuroligin 4, X-linked (221933_at), score: 0.47 NMBneuromedin B (205204_at), score: -0.61 NMUneuromedin U (206023_at), score: 0.64 NSBP1nucleosomal binding protein 1 (221606_s_at), score: 0.5 NSD1nuclear receptor binding SET domain protein 1 (219084_at), score: -0.59 NTRK2neurotrophic tyrosine kinase, receptor, type 2 (207152_at), score: 0.51 NUDT2nudix (nucleoside diphosphate linked moiety X)-type motif 2 (218609_s_at), score: -0.58 OLFM1olfactomedin 1 (213131_at), score: 0.44 OR10C1olfactory receptor, family 10, subfamily C, member 1 (221339_at), score: 0.55 OR2S2olfactory receptor, family 2, subfamily S, member 2 (221409_at), score: 0.68 ORC1Lorigin recognition complex, subunit 1-like (yeast) (205085_at), score: 0.45 PAN2PAN2 poly(A) specific ribonuclease subunit homolog (S. cerevisiae) (203117_s_at), score: -0.65 PCCApropionyl Coenzyme A carboxylase, alpha polypeptide (203860_at), score: -0.58 PCDH11Yprotocadherin 11 Y-linked (211227_s_at), score: 0.67 PDE3Aphosphodiesterase 3A, cGMP-inhibited (206388_at), score: 0.79 PEG3paternally expressed 3 (209242_at), score: 0.65 PGPEP1pyroglutamyl-peptidase I (219891_at), score: -0.65 PIAS4protein inhibitor of activated STAT, 4 (212881_at), score: -0.57 PIGNphosphatidylinositol glycan anchor biosynthesis, class N (219048_at), score: 0.44 PLEKHA9pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 9 (220157_x_at), score: -0.59 PLS1plastin 1 (I isoform) (205190_at), score: 0.73 PLXNB3plexin B3 (205957_at), score: 0.53 POU4F1POU class 4 homeobox 1 (206940_s_at), score: 0.49 PPIAL4Apeptidylprolyl isomerase A (cyclophilin A)-like 4A (217136_at), score: 0.63 PRKAB2protein kinase, AMP-activated, beta 2 non-catalytic subunit (214474_at), score: -0.56 PRKG2protein kinase, cGMP-dependent, type II (207505_at), score: 0.61 PRLprolactin (205445_at), score: 0.74 PROL1proline rich, lacrimal 1 (208004_at), score: 0.55 PRR3proline rich 3 (204795_at), score: -0.56 PRSS3protease, serine, 3 (207463_x_at), score: 0.56 PTCD1pentatricopeptide repeat domain 1 (218956_s_at), score: -0.69 PTP4A3protein tyrosine phosphatase type IVA, member 3 (209695_at), score: 0.45 R3HDM2R3H domain containing 2 (203831_at), score: -0.75 RAB30RAB30, member RAS oncogene family (206530_at), score: -0.59 RAB40BRAB40B, member RAS oncogene family (217597_x_at), score: -0.63 RECQL5RecQ protein-like 5 (34063_at), score: 0.69 RHBDF2rhomboid 5 homolog 2 (Drosophila) (219202_at), score: 0.45 RIPK1receptor (TNFRSF)-interacting serine-threonine kinase 1 (209941_at), score: 0.47 RMND5Arequired for meiotic nuclear division 5 homolog A (S. cerevisiae) (212479_s_at), score: -0.59 RNASE2ribonuclease, RNase A family, 2 (liver, eosinophil-derived neurotoxin) (206111_at), score: 0.59 RNF2ring finger protein 2 (205215_at), score: 0.48 RP11-35N6.1plasticity related gene 3 (219732_at), score: 0.68 RP4-692D3.1hypothetical protein LOC728621 (221171_at), score: 0.45 RPIAribose 5-phosphate isomerase A (212973_at), score: -0.64 RPS6KA6ribosomal protein S6 kinase, 90kDa, polypeptide 6 (220738_s_at), score: 0.48 SAT1spermidine/spermine N1-acetyltransferase 1 (213988_s_at), score: 0.49 SCARF1scavenger receptor class F, member 1 (206995_x_at), score: 0.61 SCN1Bsodium channel, voltage-gated, type I, beta (205508_at), score: -0.64 SEMA4Fsema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4F (210124_x_at), score: -0.56 SERGEFsecretion regulating guanine nucleotide exchange factor (220482_s_at), score: -0.56 SERPINB9serpin peptidase inhibitor, clade B (ovalbumin), member 9 (209723_at), score: -0.65 SETMARSET domain and mariner transposase fusion gene (206554_x_at), score: -0.57 SF4splicing factor 4 (215004_s_at), score: -0.72 SIGLEC6sialic acid binding Ig-like lectin 6 (210796_x_at), score: 0.61 SIK2salt-inducible kinase 2 (213221_s_at), score: 0.5 SLC12A3solute carrier family 12 (sodium/chloride transporters), member 3 (215274_at), score: 0.52 SLC12A9solute carrier family 12 (potassium/chloride transporters), member 9 (220371_s_at), score: -0.64 SLC16A5solute carrier family 16, member 5 (monocarboxylic acid transporter 6) (206600_s_at), score: -0.68 SLC16A6solute carrier family 16, member 6 (monocarboxylic acid transporter 7) (207038_at), score: 0.7 SLC17A7solute carrier family 17 (sodium-dependent inorganic phosphate cotransporter), member 7 (204230_s_at), score: 0.49 SLC24A6solute carrier family 24 (sodium/potassium/calcium exchanger), member 6 (218749_s_at), score: 0.52 SLC2A1solute carrier family 2 (facilitated glucose transporter), member 1 (201250_s_at), score: -0.56 SLC30A4solute carrier family 30 (zinc transporter), member 4 (207362_at), score: 0.46 SLC38A4solute carrier family 38, member 4 (220786_s_at), score: -0.56 SLC4A10solute carrier family 4, sodium bicarbonate transporter, member 10 (206830_at), score: 0.45 SLC6A14solute carrier family 6 (amino acid transporter), member 14 (219795_at), score: 0.72 SNORA21small nucleolar RNA, H/ACA box 21 (215224_at), score: -0.7 SOX17SRY (sex determining region Y)-box 17 (219993_at), score: 0.48 SPAG16sperm associated antigen 16 (219109_at), score: -0.6 SPAM1sperm adhesion molecule 1 (PH-20 hyaluronidase, zona pellucida binding) (216989_at), score: 0.69 SPTA1spectrin, alpha, erythrocytic 1 (elliptocytosis 2) (206937_at), score: 0.69 SUPV3L1suppressor of var1, 3-like 1 (S. cerevisiae) (212894_at), score: -0.72 SYCP2synaptonemal complex protein 2 (206546_at), score: 0.49 SYNMsynemin, intermediate filament protein (212730_at), score: -0.6 SYT11synaptotagmin XI (209198_s_at), score: -0.58 TAAR2trace amine associated receptor 2 (221394_at), score: 0.84 TAF15TAF15 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 68kDa (202840_at), score: -0.58 THOC6THO complex 6 homolog (Drosophila) (218848_at), score: -0.78 THUMPD1THUMP domain containing 1 (206555_s_at), score: -0.55 TLR1toll-like receptor 1 (210176_at), score: 0.6 TMSB15Athymosin beta 15a (205347_s_at), score: -0.56 TPK1thiamin pyrophosphokinase 1 (221218_s_at), score: 0.51 TRIB2tribbles homolog 2 (Drosophila) (202478_at), score: -0.59 TRIM34tripartite motif-containing 34 (221044_s_at), score: 0.44 TRIM45tripartite motif-containing 45 (219923_at), score: -0.63 TRMT61BtRNA methyltransferase 61 homolog B (S. cerevisiae) (221229_s_at), score: -0.61 TSPY1testis specific protein, Y-linked 1 (216374_at), score: 0.53 TWISTNBTWIST neighbor (214729_at), score: 0.45 UCP2uncoupling protein 2 (mitochondrial, proton carrier) (208998_at), score: -0.65 ZBTB40zinc finger and BTB domain containing 40 (203958_s_at), score: -0.66 ZHX2zinc fingers and homeoboxes 2 (203556_at), score: -0.55 ZKSCAN5zinc finger with KRAB and SCAN domains 5 (203730_s_at), score: 0.46 ZNF133zinc finger protein 133 (216960_s_at), score: -0.75 ZNF137zinc finger protein 137 (207394_at), score: 0.49 ZNF14zinc finger protein 14 (219854_at), score: -0.61 ZNF174zinc finger protein 174 (210290_at), score: -0.7 ZNF175zinc finger protein 175 (205497_at), score: 0.5 ZNF192zinc finger protein 192 (206579_at), score: 0.44 ZNF219zinc finger protein 219 (219314_s_at), score: 0.49 ZNF287zinc finger protein 287 (216710_x_at), score: 0.71 ZNF434zinc finger protein 434 (218937_at), score: -0.65 ZNF551zinc finger protein 551 (211721_s_at), score: 0.46 ZNF749zinc finger protein 749 (215289_at), score: 0.69
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-TABM-263-raw-cel-1515486011.cel | 19 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486191.cel | 28 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486391.cel | 38 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485911.cel | 14 | 6 | Cycle | hgu133a2 | none | Cycle 1 |