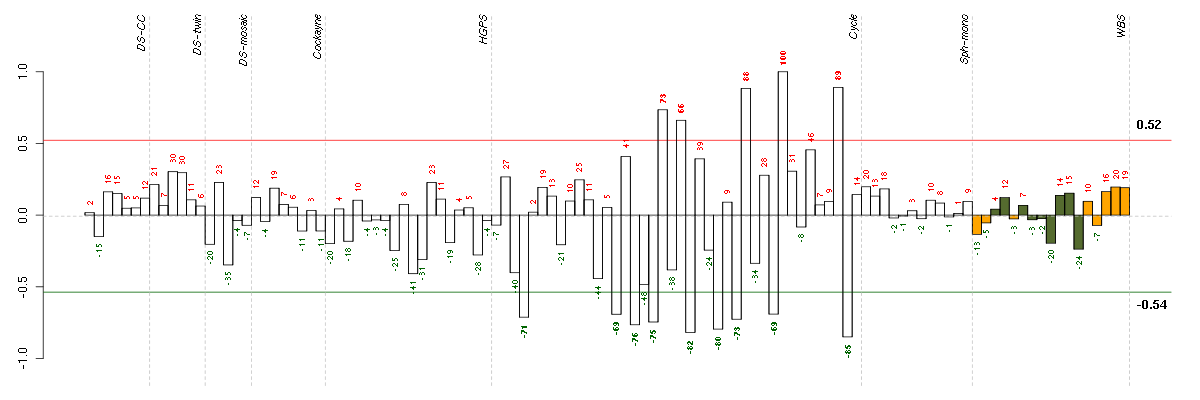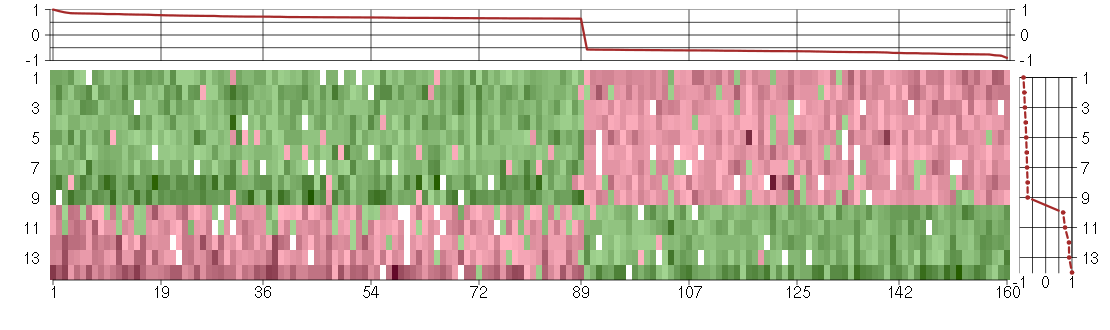

Under-expression is coded with green,
over-expression with red color.



ACANaggrecan (205679_x_at), score: 0.75 ADCK2aarF domain containing kinase 2 (221893_s_at), score: -0.8 ADRB2adrenergic, beta-2-, receptor, surface (206170_at), score: -0.67 ANXA10annexin A10 (210143_at), score: -0.72 APOBEC3Gapolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 3G (204205_at), score: 0.66 ASCC1activating signal cointegrator 1 complex subunit 1 (219336_s_at), score: 0.66 ASF1BASF1 anti-silencing function 1 homolog B (S. cerevisiae) (218115_at), score: -0.68 ASTN2astrotactin 2 (209693_at), score: 0.78 ATAD5ATPase family, AAA domain containing 5 (220223_at), score: -0.58 ATP6V0E1ATPase, H+ transporting, lysosomal 9kDa, V0 subunit e1 (214244_s_at), score: 0.82 BAT2HLA-B associated transcript 2 (212081_x_at), score: -0.59 BEST1bestrophin 1 (207671_s_at), score: 0.67 BRIP1BRCA1 interacting protein C-terminal helicase 1 (221703_at), score: -0.72 BTN3A1butyrophilin, subfamily 3, member A1 (209770_at), score: 0.77 C13orf27chromosome 13 open reading frame 27 (213346_at), score: -0.63 C19orf22chromosome 19 open reading frame 22 (221764_at), score: -0.59 C1orf66chromosome 1 open reading frame 66 (218914_at), score: 0.7 C9orf40chromosome 9 open reading frame 40 (218904_s_at), score: -0.67 CAMK1calcium/calmodulin-dependent protein kinase I (204392_at), score: -0.62 CBX4chromobox homolog 4 (Pc class homolog, Drosophila) (206724_at), score: -0.58 CCDC28Acoiled-coil domain containing 28A (209479_at), score: 0.72 CCDC94coiled-coil domain containing 94 (204335_at), score: 0.69 CCNE1cyclin E1 (213523_at), score: -0.75 CDC14BCDC14 cell division cycle 14 homolog B (S. cerevisiae) (208022_s_at), score: 0.68 CDC25Acell division cycle 25 homolog A (S. pombe) (204695_at), score: -0.6 CDC42BPBCDC42 binding protein kinase beta (DMPK-like) (217849_s_at), score: -0.62 CDCA4cell division cycle associated 4 (218399_s_at), score: -0.76 CDCP1CUB domain containing protein 1 (218451_at), score: -0.7 CDKL3cyclin-dependent kinase-like 3 (219831_at), score: 0.68 CDT1chromatin licensing and DNA replication factor 1 (209832_s_at), score: -0.71 CENPTcentromere protein T (218148_at), score: -0.67 CHPcalcium binding protein P22 (214665_s_at), score: 0.69 CHRNB2cholinergic receptor, nicotinic, beta 2 (neuronal) (206635_at), score: 0.76 CLCF1cardiotrophin-like cytokine factor 1 (219500_at), score: -0.62 CLCN6chloride channel 6 (203950_s_at), score: 0.7 CNNM2cyclin M2 (206818_s_at), score: 0.7 CORO2Bcoronin, actin binding protein, 2B (209789_at), score: -0.76 CPA4carboxypeptidase A4 (205832_at), score: 0.81 CRYBG3beta-gamma crystallin domain containing 3 (214030_at), score: 0.72 CTNScystinosis, nephropathic (36566_at), score: 0.67 CTSScathepsin S (202901_x_at), score: -0.63 CTTNcortactin (201059_at), score: 0.73 DCAKDdephospho-CoA kinase domain containing (221224_s_at), score: 0.71 DCUN1D2DCN1, defective in cullin neddylation 1, domain containing 2 (S. cerevisiae) (219116_s_at), score: 0.65 DDIT3DNA-damage-inducible transcript 3 (209383_at), score: 0.68 DHRS2dehydrogenase/reductase (SDR family) member 2 (214079_at), score: -0.62 DIRAS3DIRAS family, GTP-binding RAS-like 3 (215506_s_at), score: -0.67 DOK4docking protein 4 (209691_s_at), score: -0.73 DPYSL2dihydropyrimidinase-like 2 (200762_at), score: 0.64 DSN1DSN1, MIND kinetochore complex component, homolog (S. cerevisiae) (219512_at), score: -0.74 DSPPdentin sialophosphoprotein (221681_s_at), score: 0.65 DUSP5dual specificity phosphatase 5 (209457_at), score: -0.64 DVL2dishevelled, dsh homolog 2 (Drosophila) (57532_at), score: 0.67 EIF4Beukaryotic translation initiation factor 4B (219599_at), score: 0.66 ELNelastin (212670_at), score: -0.63 EVLEnah/Vasp-like (217838_s_at), score: -0.6 FAM117Afamily with sequence similarity 117, member A (221249_s_at), score: 0.78 FAM29Afamily with sequence similarity 29, member A (218602_s_at), score: -0.65 FAM63Bfamily with sequence similarity 63, member B (214691_x_at), score: -0.57 FLJ12529pre-mRNA cleavage factor I, 59 kDa subunit (217866_at), score: -0.61 FOSL1FOS-like antigen 1 (204420_at), score: -0.58 FOXK2forkhead box K2 (203064_s_at), score: -0.63 FZD4frizzled homolog 4 (Drosophila) (218665_at), score: 0.7 GAL3ST4galactose-3-O-sulfotransferase 4 (219815_at), score: 0.75 GALNSgalactosamine (N-acetyl)-6-sulfate sulfatase (206335_at), score: 0.79 GAS1growth arrest-specific 1 (204457_s_at), score: 0.66 GPR144G protein-coupled receptor 144 (216289_at), score: 0.73 GYPCglycophorin C (Gerbich blood group) (202947_s_at), score: 0.69 HAB1B1 for mucin (215778_x_at), score: 0.65 HINFPhistone H4 transcription factor (206495_s_at), score: 0.84 HMGA1high mobility group AT-hook 1 (210457_x_at), score: -0.64 IFT140intraflagellar transport 140 homolog (Chlamydomonas) (204792_s_at), score: 0.67 IKZF4IKAROS family zinc finger 4 (Eos) (208472_at), score: 0.67 IL13RA2interleukin 13 receptor, alpha 2 (206172_at), score: -0.6 IL33interleukin 33 (209821_at), score: -0.7 ING4inhibitor of growth family, member 4 (48825_at), score: 0.73 ITGALintegrin, alpha L (antigen CD11A (p180), lymphocyte function-associated antigen 1; alpha polypeptide) (213475_s_at), score: 0.84 KRT33Akeratin 33A (208483_x_at), score: -0.71 LOC388152hypothetical LOC388152 (220602_s_at), score: 0.85 MAP1Smicrotubule-associated protein 1S (218522_s_at), score: -0.81 MAPRE3microtubule-associated protein, RP/EB family, member 3 (203842_s_at), score: 0.65 MARCH8membrane-associated ring finger (C3HC4) 8 (221824_s_at), score: 0.7 MGAMAX gene associated (212945_s_at), score: 0.64 MGAT1mannosyl (alpha-1,3-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase (201126_s_at), score: -0.57 MMP16matrix metallopeptidase 16 (membrane-inserted) (207012_at), score: -0.6 MOSPD1motile sperm domain containing 1 (218853_s_at), score: 0.72 MTMR1myotubularin related protein 1 (214975_s_at), score: -0.64 MUC1mucin 1, cell surface associated (207847_s_at), score: -0.59 NEDD4Lneural precursor cell expressed, developmentally down-regulated 4-like (212445_s_at), score: -0.63 NFATC2IPnuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 2 interacting protein (217526_at), score: -0.75 NFIXnuclear factor I/X (CCAAT-binding transcription factor) (209807_s_at), score: 0.8 NTHL1nth endonuclease III-like 1 (E. coli) (209731_at), score: 0.72 ORAI3ORAI calcium release-activated calcium modulator 3 (221864_at), score: 0.65 PCCApropionyl Coenzyme A carboxylase, alpha polypeptide (203860_at), score: 0.82 PGPEP1pyroglutamyl-peptidase I (219891_at), score: 0.75 PIP5K1Cphosphatidylinositol-4-phosphate 5-kinase, type I, gamma (212518_at), score: -0.61 PKIAprotein kinase (cAMP-dependent, catalytic) inhibitor alpha (204612_at), score: -0.66 PLAURplasminogen activator, urokinase receptor (211924_s_at), score: -0.62 POM121POM121 membrane glycoprotein (rat) (212178_s_at), score: -0.66 POU6F1POU class 6 homeobox 1 (205878_at), score: 0.69 PRKG2protein kinase, cGMP-dependent, type II (207505_at), score: -0.6 PRSS3protease, serine, 3 (207463_x_at), score: -0.9 PTCD1pentatricopeptide repeat domain 1 (218956_s_at), score: 0.75 PXNpaxillin (211823_s_at), score: -0.74 RAB13RAB13, member RAS oncogene family (202252_at), score: 0.69 RAD54L2RAD54-like 2 (S. cerevisiae) (213205_s_at), score: -0.6 RBMS3RNA binding motif, single stranded interacting protein (206767_at), score: 0.72 RDH11retinol dehydrogenase 11 (all-trans/9-cis/11-cis) (217775_s_at), score: 0.67 RHOBTB1Rho-related BTB domain containing 1 (212651_at), score: 0.81 RIPK1receptor (TNFRSF)-interacting serine-threonine kinase 1 (209941_at), score: -0.75 RPL18AP6ribosomal protein L18a pseudogene 6 (216383_at), score: 0.67 RPL27Aribosomal protein L27a (212044_s_at), score: 0.89 RPL38ribosomal protein L38 (202028_s_at), score: 0.8 RPLP2ribosomal protein, large, P2 (200908_s_at), score: 0.65 RPS11ribosomal protein S11 (213350_at), score: 0.71 RPS6KA4ribosomal protein S6 kinase, 90kDa, polypeptide 4 (204632_at), score: -0.59 RRAGBRas-related GTP binding B (205540_s_at), score: 0.65 SAT1spermidine/spermine N1-acetyltransferase 1 (213988_s_at), score: -0.76 SBF1SET binding factor 1 (39835_at), score: -0.61 SCDstearoyl-CoA desaturase (delta-9-desaturase) (200832_s_at), score: 0.67 SERPINB2serpin peptidase inhibitor, clade B (ovalbumin), member 2 (204614_at), score: -0.65 SETMARSET domain and mariner transposase fusion gene (206554_x_at), score: 0.84 SLC16A5solute carrier family 16, member 5 (monocarboxylic acid transporter 6) (206600_s_at), score: 0.76 SLC16A6solute carrier family 16, member 6 (monocarboxylic acid transporter 7) (207038_at), score: -0.64 SLC20A1solute carrier family 20 (phosphate transporter), member 1 (201920_at), score: -0.58 SLC25A28solute carrier family 25, member 28 (221432_s_at), score: -0.69 SLC29A1solute carrier family 29 (nucleoside transporters), member 1 (201801_s_at), score: -0.76 SNHG3small nucleolar RNA host gene 3 (non-protein coding) (215011_at), score: 0.66 SNORA21small nucleolar RNA, H/ACA box 21 (215224_at), score: 0.94 SNRPEsmall nuclear ribonucleoprotein polypeptide E (215450_at), score: 0.64 SOLHsmall optic lobes homolog (Drosophila) (204275_at), score: -0.61 SPRY4sprouty homolog 4 (Drosophila) (221489_s_at), score: -0.68 SSR4signal sequence receptor, delta (translocon-associated protein delta) (201004_at), score: 0.67 STAT2signal transducer and activator of transcription 2, 113kDa (205170_at), score: 0.65 SUPV3L1suppressor of var1, 3-like 1 (S. cerevisiae) (212894_at), score: 0.82 SYT1synaptotagmin I (203999_at), score: -0.66 SYT11synaptotagmin XI (209198_s_at), score: 0.85 TAL1T-cell acute lymphocytic leukemia 1 (206283_s_at), score: 0.69 TMEM22transmembrane protein 22 (219569_s_at), score: -0.59 TMSB4Ythymosin beta 4, Y-linked (206769_at), score: 0.66 TP53TG1TP53 target 1 (non-protein coding) (210241_s_at), score: 1 TRIM2tripartite motif-containing 2 (202341_s_at), score: 0.7 TXNthioredoxin (216609_at), score: 0.83 TYMPthymidine phosphorylase (217497_at), score: 0.64 TYRO3TYRO3 protein tyrosine kinase (211432_s_at), score: -0.63 UBE4Bubiquitination factor E4B (UFD2 homolog, yeast) (202317_s_at), score: 0.66 UBR7ubiquitin protein ligase E3 component n-recognin 7 (putative) (218108_at), score: -0.59 USP47ubiquitin specific peptidase 47 (221518_s_at), score: 0.7 VAMP1vesicle-associated membrane protein 1 (synaptobrevin 1) (213326_at), score: 0.65 VCPIP1valosin containing protein (p97)/p47 complex interacting protein 1 (219810_at), score: 0.69 VEGFBvascular endothelial growth factor B (203683_s_at), score: -0.67 VWA5Avon Willebrand factor A domain containing 5A (210102_at), score: 0.66 WDTC1WD and tetratricopeptide repeats 1 (215497_s_at), score: 0.69 WFDC8WAP four-disulfide core domain 8 (215276_at), score: 0.72 ZDHHC18zinc finger, DHHC-type containing 18 (212860_at), score: -0.71 ZFYVE26zinc finger, FYVE domain containing 26 (37943_at), score: 0.77 ZHX2zinc fingers and homeoboxes 2 (203556_at), score: 0.7 ZNF281zinc finger protein 281 (218401_s_at), score: 0.65 ZNF318zinc finger protein 318 (203521_s_at), score: -0.58 ZNF516zinc finger protein 516 (203604_at), score: 0.76
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-TABM-263-raw-cel-1515486411.cel | 39 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486071.cel | 22 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486131.cel | 25 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485951.cel | 16 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485991.cel | 18 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486171.cel | 27 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485711.cel | 4 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485911.cel | 14 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486251.cel | 31 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486051.cel | 21 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486011.cel | 19 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486191.cel | 28 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486391.cel | 38 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486271.cel | 32 | 6 | Cycle | hgu133a2 | none | Cycle 1 |