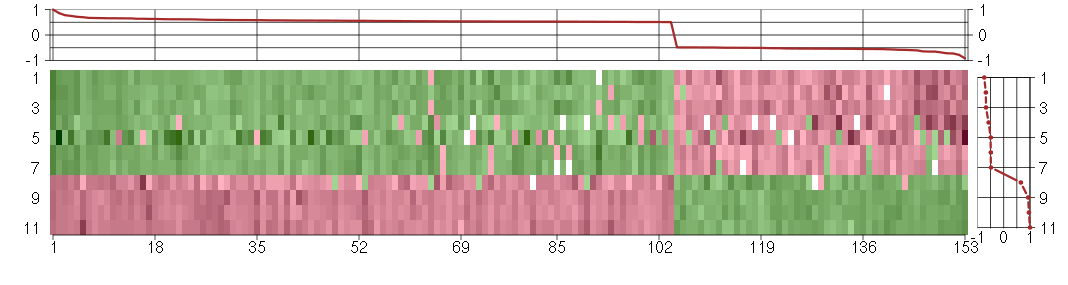

Under-expression is coded with green,
over-expression with red color.

cell morphogenesis
The developmental process by which the size or shape of a cell is generated and organized. Morphogenesis pertains to the creation of form.
cell morphogenesis involved in differentiation
The change in form (cell shape and size) that occurs when relatively unspecialized cells, e.g. embryonic or regenerative cells, acquire specialized structural and/or functional features that characterize the cells, tissues, or organs of the mature organism or some other relatively stable phase of the organism's life history.
cell motion
Any process involved in the controlled movement of a cell.
multicellular organismal development
The biological process whose specific outcome is the progression of a multicellular organism over time from an initial condition (e.g. a zygote or a young adult) to a later condition (e.g. a multicellular animal or an aged adult).
anatomical structure morphogenesis
The process by which anatomical structures are generated and organized. Morphogenesis pertains to the creation of form.
nervous system development
The process whose specific outcome is the progression of nervous tissue over time, from its formation to its mature state.
axonogenesis
Generation of a long process of a neuron, that carries efferent (outgoing) action potentials from the cell body towards target cells.
axon guidance
The process by which the migration of an axon growth cone is directed to a specific target site in response to a combination of attractive and repulsive cues.
biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
cellular process
Any process that is carried out at the cellular level, but not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
cellular component organization
A process that is carried out at the cellular level which results in the formation, arrangement of constituent parts, or disassembly of a cellular component.
neurogenesis
Generation of cells within the nervous system.
cell projection organization
A process that is carried out at the cellular level which results in the formation, arrangement of constituent parts, or disassembly of a prolongation or process extending from a cell, e.g. a flagellum or axon.
cell differentiation
The process whereby relatively unspecialized cells, e.g. embryonic or regenerative cells, acquire specialized structural and/or functional features that characterize the cells, tissues, or organs of the mature organism or some other relatively stable phase of the organism's life history. Differentiation includes the processes involved in commitment of a cell to a specific fate.
neuron differentiation
The process whereby a relatively unspecialized cell acquires specialized features of a neuron.
neurite development
The process whose specific outcome is the progression of the neurite over time, from its formation to the mature structure. The neurite is any process extending from a neural cell, such as axons or dendrites.
multicellular organismal process
Any biological process, occurring at the level of a multicellular organism, pertinent to its function.
developmental process
A biological process whose specific outcome is the progression of an integrated living unit: an anatomical structure (which may be a subcellular structure, cell, tissue, or organ), or organism over time from an initial condition to a later condition.
cellular structure morphogenesis
The process by which cellular structures are generated and organized. Morphogenesis pertains to the creation of form.
cell part morphogenesis
The process by which the anatomical structures of a cell part are generated and organized. Morphogenesis pertains to the creation of form.
cell development
The process whose specific outcome is the progression of the cell over time, from its formation to the mature structure. Cell development does not include the steps involved in committing a cell to a specific fate.
neuron development
The process whose specific outcome is the progression of a neuron over time, from initial commitment of the cell to a specific fate, to the fully functional differentiated cell.
cell morphogenesis involved in neuron differentiation
The process by which the structures of a neuron are generated and organized. This process occurs while the initially relatively unspecialized cell is acquiring the specialized features of a neuron.
generation of neurons
The process by which nerve cells are generated. This includes the production of neuroblasts and their differentiation into neurons.
system development
The process whose specific outcome is the progression of an organismal system over time, from its formation to the mature structure. A system is a regularly interacting or interdependent group of organs or tissues that work together to carry out a given biological process.
neurite morphogenesis
The process by which the anatomical structures of neurites are generated and organized. Morphogenesis pertains to the creation of form. A neurite is any process extending from a neural cell, such as axons or dendrites.
anatomical structure development
The biological process whose specific outcome is the progression of an anatomical structure from an initial condition to its mature state. This process begins with the formation of the structure and ends with the mature structure, whatever form that may be including its natural destruction. An anatomical structure is any biological entity that occupies space and is distinguished from its surroundings. Anatomical structures can be macroscopic such as a carpel, or microscopic such as an acrosome.
cell projection morphogenesis
The process by which the anatomical structures of a cell projection are generated and organized. Morphogenesis pertains to the creation of form.
cellular developmental process
A biological process whose specific outcome is the progression of a cell over time from an initial condition to a later condition.
localization
Any process by which a cell, a substance, or a cellular entity, such as a protein complex or organelle, is transported to, and/or maintained in a specific location.
localization of cell
Any process by which a cell is transported to, and/or maintained in, a specific location.
all
This term is the most general term possible
multicellular organismal development
The biological process whose specific outcome is the progression of a multicellular organism over time from an initial condition (e.g. a zygote or a young adult) to a later condition (e.g. a multicellular animal or an aged adult).
cellular developmental process
A biological process whose specific outcome is the progression of a cell over time from an initial condition to a later condition.
cellular structure morphogenesis
The process by which cellular structures are generated and organized. Morphogenesis pertains to the creation of form.
cellular structure morphogenesis
The process by which cellular structures are generated and organized. Morphogenesis pertains to the creation of form.
anatomical structure morphogenesis
The process by which anatomical structures are generated and organized. Morphogenesis pertains to the creation of form.
system development
The process whose specific outcome is the progression of an organismal system over time, from its formation to the mature structure. A system is a regularly interacting or interdependent group of organs or tissues that work together to carry out a given biological process.
cell motion
Any process involved in the controlled movement of a cell.
cell morphogenesis involved in neuron differentiation
The process by which the structures of a neuron are generated and organized. This process occurs while the initially relatively unspecialized cell is acquiring the specialized features of a neuron.
cell development
The process whose specific outcome is the progression of the cell over time, from its formation to the mature structure. Cell development does not include the steps involved in committing a cell to a specific fate.
cell morphogenesis involved in differentiation
The change in form (cell shape and size) that occurs when relatively unspecialized cells, e.g. embryonic or regenerative cells, acquire specialized structural and/or functional features that characterize the cells, tissues, or organs of the mature organism or some other relatively stable phase of the organism's life history.
cell projection morphogenesis
The process by which the anatomical structures of a cell projection are generated and organized. Morphogenesis pertains to the creation of form.
neuron differentiation
The process whereby a relatively unspecialized cell acquires specialized features of a neuron.
neurite morphogenesis
The process by which the anatomical structures of neurites are generated and organized. Morphogenesis pertains to the creation of form. A neurite is any process extending from a neural cell, such as axons or dendrites.
neuron development
The process whose specific outcome is the progression of a neuron over time, from initial commitment of the cell to a specific fate, to the fully functional differentiated cell.
neurogenesis
Generation of cells within the nervous system.
neurite development
The process whose specific outcome is the progression of the neurite over time, from its formation to the mature structure. The neurite is any process extending from a neural cell, such as axons or dendrites.
cell projection morphogenesis
The process by which the anatomical structures of a cell projection are generated and organized. Morphogenesis pertains to the creation of form.
neurite morphogenesis
The process by which the anatomical structures of neurites are generated and organized. Morphogenesis pertains to the creation of form. A neurite is any process extending from a neural cell, such as axons or dendrites.
axon guidance
The process by which the migration of an axon growth cone is directed to a specific target site in response to a combination of attractive and repulsive cues.


molecular_function
Elemental activities, such as catalysis or binding, describing the actions of a gene product at the molecular level. A given gene product may exhibit one or more molecular functions.
catalytic activity
Catalysis of a biochemical reaction at physiological temperatures. In biologically catalyzed reactions, the reactants are known as substrates, and the catalysts are naturally occurring macromolecular substances known as enzymes. Enzymes possess specific binding sites for substrates, and are usually composed wholly or largely of protein, but RNA that has catalytic activity (ribozyme) is often also regarded as enzymatic.
protein kinase activity
Catalysis of the phosphorylation of an amino acid residue in a protein, usually according to the reaction: a protein + ATP = a phosphoprotein + ADP.
protein tyrosine kinase activity
Catalysis of the reaction: ATP + a protein tyrosine = ADP + protein tyrosine phosphate.
transmembrane receptor protein tyrosine kinase activity
Catalysis of the reaction: ATP + a protein-L-tyrosine = ADP + a protein-L-tyrosine phosphate, to initiate a change in cell activity.
signal transducer activity
Mediates the transfer of a signal from the outside to the inside of a cell by means other than the introduction of the signal molecule itself into the cell.
receptor activity
Combining with an extracellular or intracellular messenger to initiate a change in cell activity.
transmembrane receptor activity
Combining with an extracellular or intracellular messenger to initiate a change in cell activity, and spanning to the membrane of either the cell or an organelle.
vascular endothelial growth factor receptor activity
Combining with vascular endothelial growth factor to initiate a change in cell activity.
kinase activity
Catalysis of the transfer of a phosphate group, usually from ATP, to a substrate molecule.
transferase activity
Catalysis of the transfer of a group, e.g. a methyl group, glycosyl group, acyl group, phosphorus-containing, or other groups, from one compound (generally regarded as the donor) to another compound (generally regarded as the acceptor). Transferase is the systematic name for any enzyme of EC class 2.
transferase activity, transferring phosphorus-containing groups
Catalysis of the transfer of a phosphorus-containing group from one compound (donor) to another (acceptor).
phosphotransferase activity, alcohol group as acceptor
Catalysis of the transfer of a phosphorus-containing group from one compound (donor) to an alcohol group (acceptor).
transmembrane receptor protein kinase activity
NA
molecular transducer activity
The molecular function that accepts an input of one form and creates an output of a different form.
all
This term is the most general term possible
protein kinase activity
Catalysis of the phosphorylation of an amino acid residue in a protein, usually according to the reaction: a protein + ATP = a phosphoprotein + ADP.
transmembrane receptor protein kinase activity
NA
transmembrane receptor protein tyrosine kinase activity
Catalysis of the reaction: ATP + a protein-L-tyrosine = ADP + a protein-L-tyrosine phosphate, to initiate a change in cell activity.
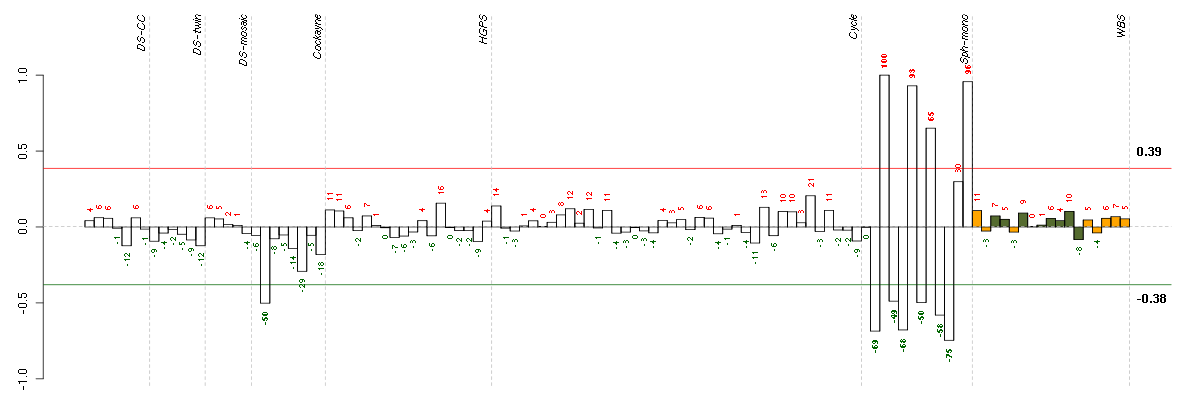
ABLIM1actin binding LIM protein 1 (200965_s_at), score: 0.53 AEBP1AE binding protein 1 (201792_at), score: 0.62 AKAP12A kinase (PRKA) anchor protein 12 (210517_s_at), score: 0.57 AKAP2A kinase (PRKA) anchor protein 2 (202759_s_at), score: 0.57 AKR1B10aldo-keto reductase family 1, member B10 (aldose reductase) (206561_s_at), score: -0.65 ALDH1A1aldehyde dehydrogenase 1 family, member A1 (212224_at), score: 0.52 ARHGAP5Rho GTPase activating protein 5 (217936_at), score: 0.59 ARL15ADP-ribosylation factor-like 15 (219842_at), score: 0.52 ARMCX1armadillo repeat containing, X-linked 1 (218694_at), score: 0.64 BAZ2Bbromodomain adjacent to zinc finger domain, 2B (203080_s_at), score: 0.55 BMP7bone morphogenetic protein 7 (209590_at), score: -0.54 C14orf169chromosome 14 open reading frame 169 (219526_at), score: 0.65 C20orf111chromosome 20 open reading frame 111 (209020_at), score: -0.49 C8orf4chromosome 8 open reading frame 4 (218541_s_at), score: 0.54 CAND1cullin-associated and neddylation-dissociated 1 (208839_s_at), score: 0.52 CAND2cullin-associated and neddylation-dissociated 2 (putative) (213547_at), score: 0.58 CD24CD24 molecule (209771_x_at), score: -0.53 CD248CD248 molecule, endosialin (219025_at), score: 0.56 CDH6cadherin 6, type 2, K-cadherin (fetal kidney) (205532_s_at), score: -0.49 CDKN1Acyclin-dependent kinase inhibitor 1A (p21, Cip1) (202284_s_at), score: 0.57 CELSR1cadherin, EGF LAG seven-pass G-type receptor 1 (flamingo homolog, Drosophila) (41660_at), score: -0.55 CELSR3cadherin, EGF LAG seven-pass G-type receptor 3 (flamingo homolog, Drosophila) (40020_at), score: -0.78 CHMP1Achromatin modifying protein 1A (201933_at), score: -0.54 CLEC11AC-type lectin domain family 11, member A (211709_s_at), score: 0.55 CLEC2BC-type lectin domain family 2, member B (209732_at), score: -0.55 COL3A1collagen, type III, alpha 1 (215076_s_at), score: 0.67 CUGBP2CUG triplet repeat, RNA binding protein 2 (202158_s_at), score: 0.55 CYB5R2cytochrome b5 reductase 2 (220230_s_at), score: 0.53 DCNdecorin (211896_s_at), score: 0.57 DDAH1dimethylarginine dimethylaminohydrolase 1 (209094_at), score: 0.54 DERL1Der1-like domain family, member 1 (219402_s_at), score: -0.5 DGKDdiacylglycerol kinase, delta 130kDa (208072_s_at), score: -0.68 DNAJC7DnaJ (Hsp40) homolog, subfamily C, member 7 (202416_at), score: -0.5 DRG1developmentally regulated GTP binding protein 1 (202810_at), score: -0.52 DSEdermatan sulfate epimerase (218854_at), score: 0.55 ELOVL4elongation of very long chain fatty acids (FEN1/Elo2, SUR4/Elo3, yeast)-like 4 (219532_at), score: 0.52 ENOSF1enolase superfamily member 1 (204142_at), score: 0.51 ENPP2ectonucleotide pyrophosphatase/phosphodiesterase 2 (209392_at), score: 0.53 EPB41L1erythrocyte membrane protein band 4.1-like 1 (212339_at), score: -0.49 EPHX1epoxide hydrolase 1, microsomal (xenobiotic) (202017_at), score: -0.55 FAM105Afamily with sequence similarity 105, member A (219694_at), score: -0.49 FAM174Bfamily with sequence similarity 174, member B (51158_at), score: 0.7 FAM65Bfamily with sequence similarity 65, member B (209829_at), score: 0.59 FCGR2AFc fragment of IgG, low affinity IIa, receptor (CD32) (203561_at), score: -0.54 FLJ10357hypothetical protein FLJ10357 (58780_s_at), score: 0.53 FLT1fms-related tyrosine kinase 1 (vascular endothelial growth factor/vascular permeability factor receptor) (222033_s_at), score: 0.6 FOXG1forkhead box G1 (206018_at), score: -0.59 FOXL2forkhead box L2 (220102_at), score: -0.53 GAL3ST4galactose-3-O-sulfotransferase 4 (219815_at), score: 0.6 GALCgalactosylceramidase (204417_at), score: 0.66 GAS1growth arrest-specific 1 (204457_s_at), score: 0.55 GPRC5BG protein-coupled receptor, family C, group 5, member B (203632_s_at), score: -0.53 GREM1gremlin 1, cysteine knot superfamily, homolog (Xenopus laevis) (218469_at), score: 1 HEY1hairy/enhancer-of-split related with YRPW motif 1 (44783_s_at), score: -0.54 HLA-Bmajor histocompatibility complex, class I, B (211911_x_at), score: -0.5 HOXA10homeobox A10 (213150_at), score: 0.56 HSD17B12hydroxysteroid (17-beta) dehydrogenase 12 (217869_at), score: 0.63 HYPKHuntingtin interacting protein K (218680_x_at), score: -0.55 IGSF3immunoglobulin superfamily, member 3 (202421_at), score: -0.72 IL23Ainterleukin 23, alpha subunit p19 (211796_s_at), score: 0.58 IPWimprinted in Prader-Willi syndrome (non-protein coding) (221974_at), score: 0.62 ISL1ISL LIM homeobox 1 (206104_at), score: 0.61 JAM3junctional adhesion molecule 3 (212813_at), score: 0.52 KYNUkynureninase (L-kynurenine hydrolase) (217388_s_at), score: -0.92 LEF1lymphoid enhancer-binding factor 1 (221558_s_at), score: -0.72 LMO7LIM domain 7 (202674_s_at), score: 0.53 LOC728855hypothetical LOC728855 (222001_x_at), score: -0.54 LPPR4plasticity related gene 1 (213496_at), score: 0.59 LRRC2leucine rich repeat containing 2 (219949_at), score: 0.53 LTBP2latent transforming growth factor beta binding protein 2 (204682_at), score: 0.58 LUMlumican (201744_s_at), score: 0.86 LXNlatexin (218729_at), score: 0.56 MED23mediator complex subunit 23 (218846_at), score: 0.53 MEIS3P1Meis homeobox 3 pseudogene 1 (214077_x_at), score: 0.52 MLF1myeloid leukemia factor 1 (204784_s_at), score: -0.53 MLLT3myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 3 (204918_s_at), score: 0.59 MOXD1monooxygenase, DBH-like 1 (209708_at), score: 0.59 MPPED2metallophosphoesterase domain containing 2 (205413_at), score: -0.65 MXRA5matrix-remodelling associated 5 (209596_at), score: 0.52 MYO1Bmyosin IB (212364_at), score: 0.78 NDNnecdin homolog (mouse) (209550_at), score: 0.51 NESnestin (218678_at), score: 0.53 NPTX1neuronal pentraxin I (204684_at), score: -0.58 NRN1neuritin 1 (218625_at), score: -0.52 NRP1neuropilin 1 (212298_at), score: 0.51 NT5E5'-nucleotidase, ecto (CD73) (203939_at), score: 0.56 OLFML3olfactomedin-like 3 (218162_at), score: 0.51 PCLOpiccolo (presynaptic cytomatrix protein) (213558_at), score: -0.6 PCTK2PCTAIRE protein kinase 2 (221918_at), score: 0.53 PDGFRAplatelet-derived growth factor receptor, alpha polypeptide (203131_at), score: 0.71 PDLIM4PDZ and LIM domain 4 (214175_x_at), score: 0.54 PEX5peroxisomal biogenesis factor 5 (203244_at), score: -0.58 PFDN2prefoldin subunit 2 (218336_at), score: -0.57 PGAP1post-GPI attachment to proteins 1 (213469_at), score: 0.51 PID1phosphotyrosine interaction domain containing 1 (219093_at), score: 0.52 PLAG1pleiomorphic adenoma gene 1 (205372_at), score: 0.54 PLAGL1pleiomorphic adenoma gene-like 1 (209318_x_at), score: 0.56 PLCB1phospholipase C, beta 1 (phosphoinositide-specific) (213222_at), score: 0.58 PLCG1phospholipase C, gamma 1 (202789_at), score: -0.51 POFUT1protein O-fucosyltransferase 1 (212349_at), score: -0.5 PPIHpeptidylprolyl isomerase H (cyclophilin H) (204228_at), score: -0.49 PRR16proline rich 16 (220014_at), score: 0.62 PSMB6proteasome (prosome, macropain) subunit, beta type, 6 (208827_at), score: -0.5 PSMD5proteasome (prosome, macropain) 26S subunit, non-ATPase, 5 (203447_at), score: 0.53 PSTPIP2proline-serine-threonine phosphatase interacting protein 2 (219938_s_at), score: 0.66 PTPRDprotein tyrosine phosphatase, receptor type, D (214043_at), score: 0.64 RECKreversion-inducing-cysteine-rich protein with kazal motifs (205407_at), score: 0.62 RGS4regulator of G-protein signaling 4 (204337_at), score: 0.58 RIPK4receptor-interacting serine-threonine kinase 4 (221215_s_at), score: -0.64 RUNX1runt-related transcription factor 1 (209360_s_at), score: 0.53 S100A4S100 calcium binding protein A4 (203186_s_at), score: 0.56 SALL2sal-like 2 (Drosophila) (213283_s_at), score: 0.53 SCRIBscribbled homolog (Drosophila) (212556_at), score: 0.54 SEMA3Fsema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3F (35666_at), score: 0.51 SERPINB9serpin peptidase inhibitor, clade B (ovalbumin), member 9 (209723_at), score: 0.57 SGK1serum/glucocorticoid regulated kinase 1 (201739_at), score: 0.53 SIM1single-minded homolog 1 (Drosophila) (206876_at), score: 0.53 SLC30A1solute carrier family 30 (zinc transporter), member 1 (212907_at), score: 0.54 SLC38A4solute carrier family 38, member 4 (220786_s_at), score: 0.56 SMA4glucuronidase, beta pseudogene (215599_at), score: -0.5 SMA5glucuronidase, beta pseudogene (215043_s_at), score: -0.53 SNF8SNF8, ESCRT-II complex subunit, homolog (S. cerevisiae) (218391_at), score: -0.55 SPANXCSPANX family, member C (220217_x_at), score: -0.54 SPHARS-phase response (cyclin-related) (206272_at), score: 0.53 SSPNsarcospan (Kras oncogene-associated gene) (204963_at), score: 0.51 STAT6signal transducer and activator of transcription 6, interleukin-4 induced (201331_s_at), score: 0.53 SULF1sulfatase 1 (212353_at), score: 0.61 SYNGR1synaptogyrin 1 (210613_s_at), score: 0.65 TBC1D9TBC1 domain family, member 9 (with GRAM domain) (212956_at), score: 0.54 TBXA2Rthromboxane A2 receptor (336_at), score: 0.63 TCF12transcription factor 12 (208986_at), score: 0.52 TCF7L2transcription factor 7-like 2 (T-cell specific, HMG-box) (212761_at), score: 0.59 TEStestis derived transcript (3 LIM domains) (202720_at), score: 0.75 TGFB2transforming growth factor, beta 2 (220407_s_at), score: -0.54 THBS2thrombospondin 2 (203083_at), score: 0.67 TMEM111transmembrane protein 111 (217882_at), score: -0.56 TMEM2transmembrane protein 2 (218113_at), score: 0.52 TMEM30Atransmembrane protein 30A (217743_s_at), score: 0.52 TNS1tensin 1 (221748_s_at), score: 0.66 TOR3Atorsin family 3, member A (218459_at), score: -0.51 TPBGtrophoblast glycoprotein (203476_at), score: 0.51 TRPC3transient receptor potential cation channel, subfamily C, member 3 (210814_at), score: -0.49 TRPS1trichorhinophalangeal syndrome I (218502_s_at), score: 0.53 TSPAN6tetraspanin 6 (209108_at), score: 0.53 TSPYL5TSPY-like 5 (213122_at), score: 0.65 TUBB6tubulin, beta 6 (209191_at), score: 0.56 ULK2unc-51-like kinase 2 (C. elegans) (204062_s_at), score: 0.61 UQCRHubiquinol-cytochrome c reductase hinge protein (202233_s_at), score: -0.49 VGLL3vestigial like 3 (Drosophila) (220327_at), score: 0.56 WT1Wilms tumor 1 (206067_s_at), score: -0.49 ZHX2zinc fingers and homeoboxes 2 (203556_at), score: 0.52 ZNF215zinc finger protein 215 (220214_at), score: 0.57 ZNF518Azinc finger protein 518A (204291_at), score: 0.57
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-GEOD-4219-raw-cel-1311956614.cel | 18 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956138.cel | 4 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956321.cel | 9 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956457.cel | 14 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-3407-raw-cel-1437949579.cel | 2 | 4 | Cockayne | hgu133a | none | CSB |
| E-GEOD-4219-raw-cel-1311956398.cel | 12 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956275.cel | 8 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956418.cel | 13 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956358.cel | 10 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956824.cel | 24 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956178.cel | 6 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |