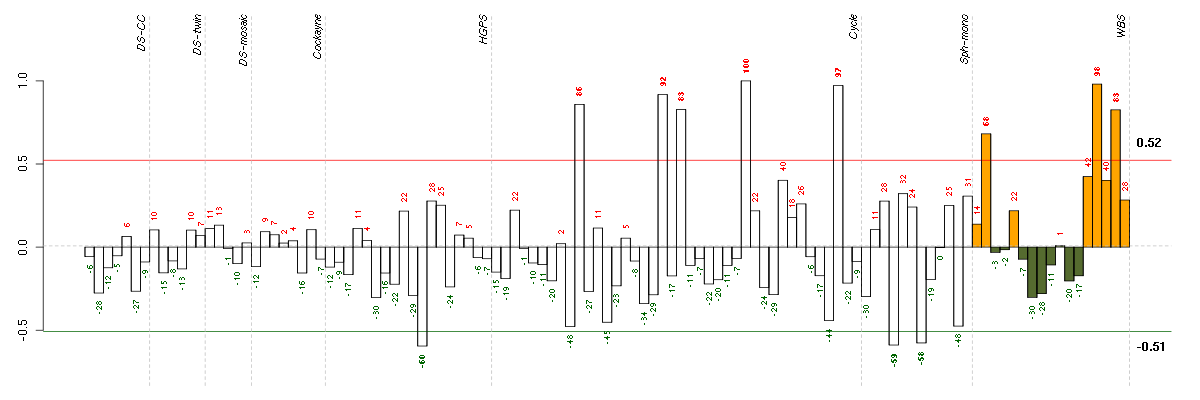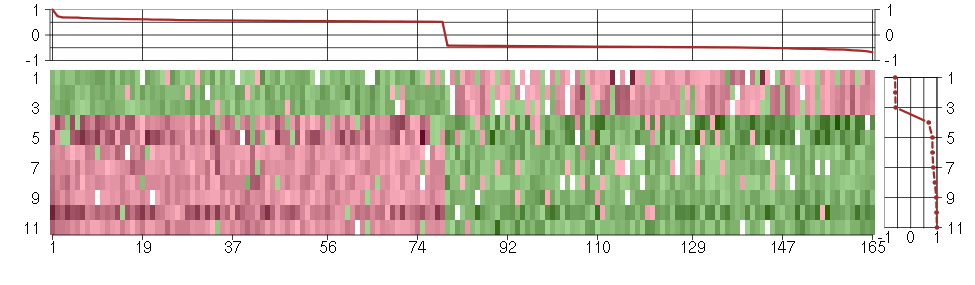

Under-expression is coded with green,
over-expression with red color.



ADAMTS6ADAM metallopeptidase with thrombospondin type 1 motif, 6 (220866_at), score: -0.55 AGBL5ATP/GTP binding protein-like 5 (218480_at), score: 0.62 AHCTF1AT hook containing transcription factor 1 (214766_s_at), score: -0.46 ALDH1A3aldehyde dehydrogenase 1 family, member A3 (203180_at), score: -0.48 ANKRD28ankyrin repeat domain 28 (213035_at), score: 0.57 ANKRD36Bankyrin repeat domain 36B (220940_at), score: -0.49 ANXA10annexin A10 (210143_at), score: -0.44 APH1Banterior pharynx defective 1 homolog B (C. elegans) (221036_s_at), score: 0.64 APOBEC3Fapolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 3F (214995_s_at), score: 0.54 ARL17ADP-ribosylation factor-like 17 (210435_at), score: -0.43 ARL2ADP-ribosylation factor-like 2 (202564_x_at), score: 0.58 ARNT2aryl-hydrocarbon receptor nuclear translocator 2 (202986_at), score: 0.55 ASB1ankyrin repeat and SOCS box-containing 1 (212819_at), score: -0.48 B3GNTL1UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase-like 1 (213589_s_at), score: 0.56 B9D1B9 protein domain 1 (210534_s_at), score: 0.62 BAXBCL2-associated X protein (211833_s_at), score: 0.55 BCCIPBRCA2 and CDKN1A interacting protein (218264_at), score: 0.55 BREbrain and reproductive organ-expressed (TNFRSF1A modulator) (205550_s_at), score: 0.53 BTDbiotinidase (204167_at), score: 0.65 BTN3A2butyrophilin, subfamily 3, member A2 (209846_s_at), score: 0.61 C14orf138chromosome 14 open reading frame 138 (218940_at), score: -0.46 C17orf39chromosome 17 open reading frame 39 (220058_at), score: 0.59 C1orf115chromosome 1 open reading frame 115 (218546_at), score: -0.42 C1orf9chromosome 1 open reading frame 9 (203429_s_at), score: -0.48 C3complement component 3 (217767_at), score: -0.5 C9orf127chromosome 9 open reading frame 127 (207839_s_at), score: 0.66 CCDC93coiled-coil domain containing 93 (209688_s_at), score: -0.43 CCNT2cyclin T2 (204645_at), score: -0.44 CD320CD320 molecule (218529_at), score: 0.6 CD55CD55 molecule, decay accelerating factor for complement (Cromer blood group) (201925_s_at), score: -0.64 CFBcomplement factor B (202357_s_at), score: -0.46 CLEC11AC-type lectin domain family 11, member A (211709_s_at), score: 0.64 CLK4CDC-like kinase 4 (210346_s_at), score: -0.42 CLUclusterin (208791_at), score: -0.68 CNPY3canopy 3 homolog (zebrafish) (217931_at), score: 0.54 CPA4carboxypeptidase A4 (205832_at), score: 0.55 CSADcysteine sulfinic acid decarboxylase (221139_s_at), score: -0.43 CSGALNACT2chondroitin sulfate N-acetylgalactosaminyltransferase 2 (222235_s_at), score: -0.43 CUTAcutA divalent cation tolerance homolog (E. coli) (221488_s_at), score: 0.52 CXCL6chemokine (C-X-C motif) ligand 6 (granulocyte chemotactic protein 2) (206336_at), score: -0.52 CYP3A43cytochrome P450, family 3, subfamily A, polypeptide 43 (211440_x_at), score: -0.42 CYTH1cytohesin 1 (202880_s_at), score: -0.46 DARS2aspartyl-tRNA synthetase 2, mitochondrial (218365_s_at), score: 0.52 DCP2DCP2 decapping enzyme homolog (S. cerevisiae) (212919_at), score: -0.5 DCPSdecapping enzyme, scavenger (218774_at), score: 0.56 DCXRdicarbonyl/L-xylulose reductase (217973_at), score: 0.55 DIRAS3DIRAS family, GTP-binding RAS-like 3 (215506_s_at), score: -0.45 DKFZP586I1420hypothetical protein DKFZp586I1420 (213546_at), score: -0.45 DPH5DPH5 homolog (S. cerevisiae) (219590_x_at), score: 0.55 DPP4dipeptidyl-peptidase 4 (211478_s_at), score: -0.47 DZIP3DAZ interacting protein 3, zinc finger (213186_at), score: 0.51 EGFL6EGF-like-domain, multiple 6 (219454_at), score: -0.61 EIF2S1eukaryotic translation initiation factor 2, subunit 1 alpha, 35kDa (201143_s_at), score: -0.52 EMP1epithelial membrane protein 1 (201325_s_at), score: -0.46 F10coagulation factor X (205620_at), score: 0.52 FAM13Bfamily with sequence similarity 13, member B (218518_at), score: -0.5 FAM149B1family with sequence similarity 149, member B1 (213896_x_at), score: 0.57 FBXO31F-box protein 31 (219785_s_at), score: 0.54 FDXRferredoxin reductase (207813_s_at), score: 0.55 FLGfilaggrin (215704_at), score: -0.49 FLJ12529pre-mRNA cleavage factor I, 59 kDa subunit (217866_at), score: -0.47 FLJ42627hypothetical LOC645644 (220352_x_at), score: -0.47 FRMD4AFERM domain containing 4A (208476_s_at), score: 0.53 GALTgalactose-1-phosphate uridylyltransferase (203179_at), score: 0.57 GARNL1GTPase activating Rap/RanGAP domain-like 1 (214855_s_at), score: -0.49 GNPDA1glucosamine-6-phosphate deaminase 1 (202382_s_at), score: 0.53 GOLGA7golgi autoantigen, golgin subfamily a, 7 (217819_at), score: -0.47 GOLSYNGolgi-localized protein (218692_at), score: -0.51 GPM6Bglycoprotein M6B (209170_s_at), score: -0.44 GPSN2glycoprotein, synaptic 2 (208336_s_at), score: 0.53 GSTM4glutathione S-transferase mu 4 (204149_s_at), score: 0.6 HCFC1R1host cell factor C1 regulator 1 (XPO1 dependent) (45714_at), score: 0.53 HERC2P2hect domain and RLD 2 pseudogene 2 (217317_s_at), score: -0.51 HISPPD2Ahistidine acid phosphatase domain containing 2A (204578_at), score: 0.55 HSPB2heat shock 27kDa protein 2 (205824_at), score: 0.57 IER2immediate early response 2 (202081_at), score: -0.42 IL13RA2interleukin 13 receptor, alpha 2 (206172_at), score: -0.43 IL33interleukin 33 (209821_at), score: -0.54 IMAASLC7A5 pseudogene (208118_x_at), score: -0.52 JHDM1Djumonji C domain containing histone demethylase 1 homolog D (S. cerevisiae) (221778_at), score: -0.48 KAL1Kallmann syndrome 1 sequence (205206_at), score: -0.49 KCTD9potassium channel tetramerisation domain containing 9 (218823_s_at), score: -0.54 KIAA0907KIAA0907 (202220_at), score: -0.45 KITv-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog (205051_s_at), score: 0.62 LMAN2Llectin, mannose-binding 2-like (221274_s_at), score: 0.58 LOC388152hypothetical LOC388152 (220602_s_at), score: 0.54 LOC729806similar to hCG1725380 (217544_at), score: -0.42 LOC791120hypothetical LOC791120 (213367_at), score: -0.44 MAP4K4mitogen-activated protein kinase kinase kinase kinase 4 (218181_s_at), score: -0.47 MEIS2Meis homeobox 2 (207480_s_at), score: 0.54 MIS12MIS12, MIND kinetochore complex component, homolog (yeast) (221559_s_at), score: -0.43 MLPHmelanophilin (218211_s_at), score: 0.74 MPImannose phosphate isomerase (202472_at), score: 0.63 MSTP9macrophage stimulating, pseudogene 9 (213382_at), score: -0.47 MTM1myotubularin 1 (36920_at), score: -0.44 NAGAN-acetylgalactosaminidase, alpha- (202943_s_at), score: 0.68 NAT13N-acetyltransferase 13 (GCN5-related) (217745_s_at), score: -0.54 NDRG4NDRG family member 4 (209159_s_at), score: 0.57 NDUFA2NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 2, 8kDa (213550_s_at), score: 0.56 NFATC2IPnuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 2 interacting protein (217526_at), score: -0.47 NIPSNAP1nipsnap homolog 1 (C. elegans) (201709_s_at), score: 0.58 NOVA1neuro-oncological ventral antigen 1 (205794_s_at), score: -0.42 NRCAMneuronal cell adhesion molecule (204105_s_at), score: -0.46 NTHL1nth endonuclease III-like 1 (E. coli) (209731_at), score: 0.68 NUDT1nudix (nucleoside diphosphate linked moiety X)-type motif 1 (204766_s_at), score: 0.52 NUPL1nucleoporin like 1 (204435_at), score: -0.58 OBFC1oligonucleotide/oligosaccharide-binding fold containing 1 (219100_at), score: 0.53 OCEL1occludin/ELL domain containing 1 (205441_at), score: 0.57 PAFAH1B3platelet-activating factor acetylhydrolase, isoform Ib, gamma subunit 29kDa (203228_at), score: 0.56 PCCApropionyl Coenzyme A carboxylase, alpha polypeptide (203860_at), score: 0.64 PCSK1proprotein convertase subtilisin/kexin type 1 (205825_at), score: -0.46 PECRperoxisomal trans-2-enoyl-CoA reductase (221142_s_at), score: 0.57 PID1phosphotyrosine interaction domain containing 1 (219093_at), score: -0.44 PIGAphosphatidylinositol glycan anchor biosynthesis, class A (205281_s_at), score: -0.53 PMS2CLPMS2 C-terminal like pseudogene (221206_at), score: -0.46 PNRC2proline-rich nuclear receptor coactivator 2 (217779_s_at), score: -0.46 PSMB9proteasome (prosome, macropain) subunit, beta type, 9 (large multifunctional peptidase 2) (204279_at), score: 0.66 PTGS1prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) (215813_s_at), score: -0.48 PYCARDPYD and CARD domain containing (221666_s_at), score: 0.65 RAB11FIP1RAB11 family interacting protein 1 (class I) (219681_s_at), score: 0.54 RAB8BRAB8B, member RAS oncogene family (219210_s_at), score: 0.56 RASSF2Ras association (RalGDS/AF-6) domain family member 2 (203185_at), score: -0.44 RBM3RNA binding motif (RNP1, RRM) protein 3 (208319_s_at), score: -0.47 RBM39RNA binding motif protein 39 (207941_s_at), score: -0.62 SAMD9sterile alpha motif domain containing 9 (219691_at), score: -0.48 SAT1spermidine/spermine N1-acetyltransferase 1 (213988_s_at), score: -0.54 SFRS12splicing factor, arginine/serine-rich 12 (212721_at), score: -0.55 SKP1S-phase kinase-associated protein 1 (200719_at), score: -0.48 SLC16A5solute carrier family 16, member 5 (monocarboxylic acid transporter 6) (206600_s_at), score: 0.63 SLC22A18solute carrier family 22, member 18 (204981_at), score: 0.55 SLC22A4solute carrier family 22 (organic cation/ergothioneine transporter), member 4 (205896_at), score: -0.43 SLC38A4solute carrier family 38, member 4 (220786_s_at), score: 0.56 SLKSTE20-like kinase (yeast) (206875_s_at), score: -0.48 SORT1sortilin 1 (212807_s_at), score: 0.54 SOX9SRY (sex determining region Y)-box 9 (202935_s_at), score: -0.57 SPA17sperm autoantigenic protein 17 (205406_s_at), score: 0.58 SPON1spondin 1, extracellular matrix protein (209436_at), score: -0.56 SS18L1synovial sarcoma translocation gene on chromosome 18-like 1 (213140_s_at), score: -0.57 STARD5StAR-related lipid transfer (START) domain containing 5 (213820_s_at), score: 0.55 STC1stanniocalcin 1 (204597_x_at), score: -0.43 SUOXsulfite oxidase (204067_at), score: 0.61 SV2Asynaptic vesicle glycoprotein 2A (203069_at), score: 0.56 SYNMsynemin, intermediate filament protein (212730_at), score: 0.52 SYT11synaptotagmin XI (209198_s_at), score: 0.69 TBC1D3TBC1 domain family, member 3 (209403_at), score: -0.45 THEM2thioesterase superfamily member 2 (204565_at), score: 0.6 THYN1thymocyte nuclear protein 1 (218491_s_at), score: 0.6 TMEM106Ctransmembrane protein 106C (201764_at), score: 0.52 TMEM87Atransmembrane protein 87A (212202_s_at), score: -0.46 TMSB15Athymosin beta 15a (205347_s_at), score: 0.69 TOXthymocyte selection-associated high mobility group box (204529_s_at), score: 0.58 TRAPPC10trafficking protein particle complex 10 (209412_at), score: -0.6 TRIM14tripartite motif-containing 14 (203148_s_at), score: 0.6 TRPV2transient receptor potential cation channel, subfamily V, member 2 (219282_s_at), score: 0.53 UBA7ubiquitin-like modifier activating enzyme 7 (203281_s_at), score: 0.54 UBXN2BUBX domain protein 2B (212934_at), score: -0.48 UCP2uncoupling protein 2 (mitochondrial, proton carrier) (208998_at), score: 1 UFM1ubiquitin-fold modifier 1 (218050_at), score: -0.57 UHRF1BP1LUHRF1 binding protein 1-like (213118_at), score: -0.42 USP12ubiquitin specific peptidase 12 (213327_s_at), score: -0.45 WHAMML1WAS protein homolog associated with actin, golgi membranes and microtubules-like 1 (213908_at), score: -0.46 YIPF1Yip1 domain family, member 1 (214733_s_at), score: 0.53 ZNF136zinc finger protein 136 (206240_s_at), score: -0.45 ZNF432zinc finger protein 432 (219848_s_at), score: -0.42 ZNF688zinc finger protein 688 (213527_s_at), score: 0.54
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-GEOD-3860-raw-cel-1561690352.cel | 11 | 5 | HGPS | hgu133a | HGPS | AG11498 |
| E-GEOD-4219-raw-cel-1311956275.cel | 8 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956398.cel | 12 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| 10590_WBS.CEL | 2 | 8 | WBS | hgu133plus2 | WBS | WBS 1 |
| F348_WBS.CEL | 16 | 8 | WBS | hgu133plus2 | WBS | WBS 1 |
| E-TABM-263-raw-cel-1515486051.cel | 21 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485831.cel | 10 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486011.cel | 19 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486391.cel | 38 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| F055_WBS.CEL | 14 | 8 | WBS | hgu133plus2 | WBS | WBS 1 |
| E-TABM-263-raw-cel-1515486191.cel | 28 | 6 | Cycle | hgu133a2 | none | Cycle 1 |