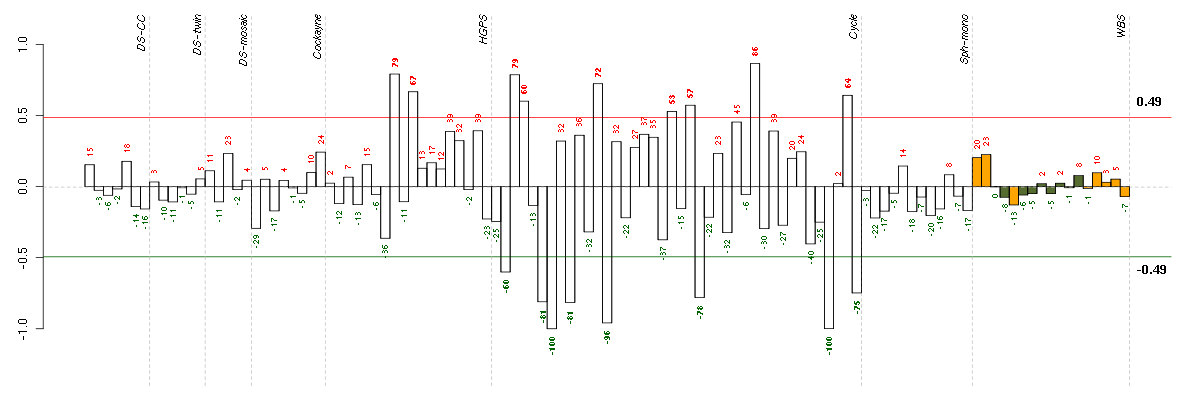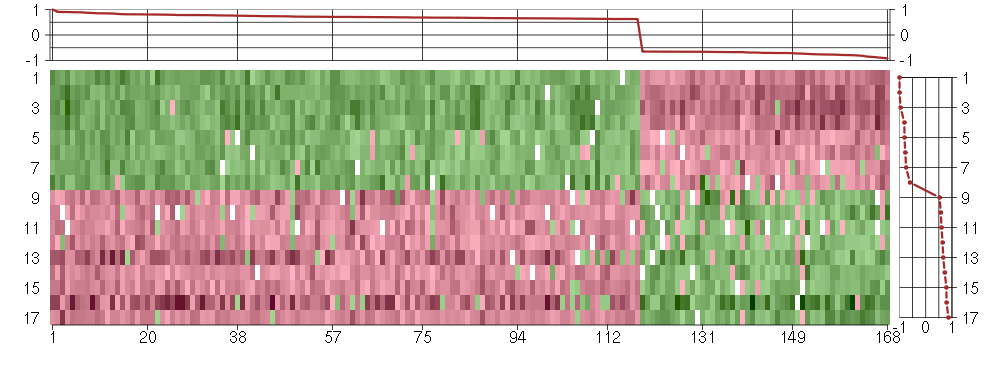

Under-expression is coded with green,
over-expression with red color.

cell adhesion
The attachment of a cell, either to another cell or to an underlying substrate such as the extracellular matrix, via cell adhesion molecules.
calcium-dependent cell-cell adhesion
The attachment of one cell to another cell via adhesion molecules that require the presence of calcium for the interaction.
biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
cellular process
Any process that is carried out at the cellular level, but not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
cell-cell adhesion
The attachment of one cell to another cell via adhesion molecules.
biological adhesion
The attachment of a cell or organism to a substrate or other organism.
all
This term is the most general term possible
cell adhesion
The attachment of a cell, either to another cell or to an underlying substrate such as the extracellular matrix, via cell adhesion molecules.


| Id | Pvalue | ExpCount | Count | Size | Term |
|---|---|---|---|---|---|
| 05050 | 6.521e-03 | 0.2362 | 4 | 14 | Dentatorubropallidoluysian atrophy (DRPLA) |
ABCD1ATP-binding cassette, sub-family D (ALD), member 1 (205142_x_at), score: 0.66 ADCK2aarF domain containing kinase 2 (221893_s_at), score: 0.81 AKAP2A kinase (PRKA) anchor protein 2 (202759_s_at), score: 0.73 ARSAarylsulfatase A (204443_at), score: 0.63 ATF5activating transcription factor 5 (204999_s_at), score: 0.63 ATN1atrophin 1 (40489_at), score: 0.8 ATP2C1ATPase, Ca++ transporting, type 2C, member 1 (211137_s_at), score: -0.71 B3GAT3beta-1,3-glucuronyltransferase 3 (glucuronosyltransferase I) (203452_at), score: 0.78 BACE2beta-site APP-cleaving enzyme 2 (217867_x_at), score: 0.71 BAIAP2BAI1-associated protein 2 (209502_s_at), score: 0.75 BAP1BRCA1 associated protein-1 (ubiquitin carboxy-terminal hydrolase) (201419_at), score: 0.65 BAT2HLA-B associated transcript 2 (212081_x_at), score: 0.78 BAT2LHLA-B associated transcript 2-like (212068_s_at), score: 0.66 BTBD2BTB (POZ) domain containing 2 (207722_s_at), score: 0.78 C14orf94chromosome 14 open reading frame 94 (218383_at), score: 0.64 C16orf57chromosome 16 open reading frame 57 (218060_s_at), score: 0.7 C19orf61chromosome 19 open reading frame 61 (221335_x_at), score: 0.68 C4orf43chromosome 4 open reading frame 43 (218513_at), score: -0.78 C5orf44chromosome 5 open reading frame 44 (218674_at), score: -0.67 CABIN1calcineurin binding protein 1 (37652_at), score: 0.65 CASC3cancer susceptibility candidate 3 (207842_s_at), score: 0.71 CASP7caspase 7, apoptosis-related cysteine peptidase (207181_s_at), score: -0.65 CDC25Bcell division cycle 25 homolog B (S. pombe) (201853_s_at), score: 0.71 CDC42BPBCDC42 binding protein kinase beta (DMPK-like) (217849_s_at), score: 0.89 CDC42EP4CDC42 effector protein (Rho GTPase binding) 4 (214721_x_at), score: 0.64 CDH13cadherin 13, H-cadherin (heart) (204726_at), score: -0.76 CENPTcentromere protein T (218148_at), score: 0.66 CHMP5chromatin modifying protein 5 (218085_at), score: -0.71 CICcapicua homolog (Drosophila) (212784_at), score: 0.85 CITcitron (rho-interacting, serine/threonine kinase 21) (212801_at), score: 0.81 CIZ1CDKN1A interacting zinc finger protein 1 (205516_x_at), score: 0.8 CNIH3cornichon homolog 3 (Drosophila) (214841_at), score: 0.71 CNOT3CCR4-NOT transcription complex, subunit 3 (203239_s_at), score: 0.87 CRTC3CREB regulated transcription coactivator 3 (218648_at), score: 0.9 CScitrate synthase (208660_at), score: 0.74 DNAJB9DnaJ (Hsp40) homolog, subfamily B, member 9 (202843_at), score: -0.77 EHBP1L1EH domain binding protein 1-like 1 (221755_at), score: 0.63 FAM18Bfamily with sequence similarity 18, member B (218446_s_at), score: -0.79 FBXL14F-box and leucine-rich repeat protein 14 (213145_at), score: 0.64 FCF1FCF1 small subunit (SSU) processome component homolog (S. cerevisiae) (219927_at), score: -0.67 FLJ12529pre-mRNA cleavage factor I, 59 kDa subunit (217866_at), score: 0.82 FOSL1FOS-like antigen 1 (204420_at), score: 0.68 FOXK2forkhead box K2 (203064_s_at), score: 0.85 FOXM1forkhead box M1 (202580_x_at), score: 0.65 G3BP2GTPase activating protein (SH3 domain) binding protein 2 (208841_s_at), score: -0.67 GABARAPL1GABA(A) receptor-associated protein like 1 (208868_s_at), score: -0.65 GGA3golgi associated, gamma adaptin ear containing, ARF binding protein 3 (209411_s_at), score: 0.68 GMCL1germ cell-less homolog 1 (Drosophila) (218458_at), score: -0.69 H1F0H1 histone family, member 0 (208886_at), score: 0.66 H1FXH1 histone family, member X (204805_s_at), score: 0.76 HDGFhepatoma-derived growth factor (high-mobility group protein 1-like) (200896_x_at), score: 0.67 HMGA1high mobility group AT-hook 1 (210457_x_at), score: 0.69 HNRNPH2heterogeneous nuclear ribonucleoprotein H2 (H') (201132_at), score: -0.79 HNRNPUL1heterogeneous nuclear ribonucleoprotein U-like 1 (209675_s_at), score: 0.76 HSD17B6hydroxysteroid (17-beta) dehydrogenase 6 homolog (mouse) (37512_at), score: -0.8 HSPA12Aheat shock 70kDa protein 12A (214434_at), score: 0.8 IDUAiduronidase, alpha-L- (205059_s_at), score: 0.79 ITGA1integrin, alpha 1 (214660_at), score: -0.7 JUPjunction plakoglobin (201015_s_at), score: 0.63 KAL1Kallmann syndrome 1 sequence (205206_at), score: -0.88 KLHL23kelch-like 23 (Drosophila) (213610_s_at), score: -0.66 LARP4La ribonucleoprotein domain family, member 4 (214155_s_at), score: -0.71 LARSleucyl-tRNA synthetase (217810_x_at), score: -0.66 LEPREL2leprecan-like 2 (204854_at), score: 0.63 LIG1ligase I, DNA, ATP-dependent (202726_at), score: 0.68 LLGL2lethal giant larvae homolog 2 (Drosophila) (203713_s_at), score: -0.66 LMF2lipase maturation factor 2 (212682_s_at), score: 0.7 LMNB2lamin B2 (216952_s_at), score: 0.65 LOC100132540similar to LOC339047 protein (214870_x_at), score: 0.68 LOC100133105hypothetical protein LOC100133105 (214237_x_at), score: -0.67 LOC339047hypothetical protein LOC339047 (221501_x_at), score: 0.72 LOC391132similar to hCG2041276 (216177_at), score: -0.77 LOC90379hypothetical protein BC002926 (221849_s_at), score: 0.66 LRDDleucine-rich repeats and death domain containing (221640_s_at), score: 0.83 LRRN3leucine rich repeat neuronal 3 (209841_s_at), score: -0.73 MAP1Smicrotubule-associated protein 1S (218522_s_at), score: 0.89 MAP3K6mitogen-activated protein kinase kinase kinase 6 (219278_at), score: 0.8 MAP7D1MAP7 domain containing 1 (217943_s_at), score: 0.75 MAPKAPK3mitogen-activated protein kinase-activated protein kinase 3 (202788_at), score: 0.71 MAST2microtubule associated serine/threonine kinase 2 (211593_s_at), score: 0.78 MAST4microtubule associated serine/threonine kinase family member 4 (40016_g_at), score: 0.63 MED15mediator complex subunit 15 (222175_s_at), score: 0.66 MFAP3Lmicrofibrillar-associated protein 3-like (205442_at), score: -0.75 MINK1misshapen-like kinase 1 (zebrafish) (214246_x_at), score: 0.68 MLL4myeloid/lymphoid or mixed-lineage leukemia 4 (203419_at), score: 0.66 MUL1mitochondrial E3 ubiquitin ligase 1 (218246_at), score: 0.71 MYO1Bmyosin IB (212364_at), score: -0.69 NBL1neuroblastoma, suppression of tumorigenicity 1 (37005_at), score: 0.84 NCKAP1NCK-associated protein 1 (217465_at), score: -0.71 NDPNorrie disease (pseudoglioma) (206022_at), score: 0.71 NDUFA5NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 5, 13kDa (201304_at), score: -0.65 NEFMneurofilament, medium polypeptide (205113_at), score: -0.9 NFATC4nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 4 (205897_at), score: 0.74 NPIPnuclear pore complex interacting protein (204538_x_at), score: 0.77 NRXN3neurexin 3 (205795_at), score: -0.66 OLFML2Bolfactomedin-like 2B (213125_at), score: 0.9 OLR1oxidized low density lipoprotein (lectin-like) receptor 1 (210004_at), score: -0.65 OR7E156Polfactory receptor, family 7, subfamily E, member 156 pseudogene (222327_x_at), score: -0.66 PAK1p21 protein (Cdc42/Rac)-activated kinase 1 (209615_s_at), score: 0.67 PARVBparvin, beta (204629_at), score: 0.77 PCDH9protocadherin 9 (219737_s_at), score: -0.73 PCDHG@protocadherin gamma cluster (215836_s_at), score: 0.68 PCDHGA1protocadherin gamma subfamily A, 1 (209079_x_at), score: 0.78 PCDHGC3protocadherin gamma subfamily C, 3 (211066_x_at), score: 0.65 PHF7PHD finger protein 7 (215622_x_at), score: -0.69 PIP5K1Cphosphatidylinositol-4-phosphate 5-kinase, type I, gamma (212518_at), score: 0.86 PKN1protein kinase N1 (202161_at), score: 0.72 PLK1polo-like kinase 1 (Drosophila) (202240_at), score: 0.7 PNPLA2patatin-like phospholipase domain containing 2 (212705_x_at), score: 0.64 POLR2Apolymerase (RNA) II (DNA directed) polypeptide A, 220kDa (202725_at), score: 0.75 POLSpolymerase (DNA directed) sigma (202466_at), score: 0.78 POM121POM121 membrane glycoprotein (rat) (212178_s_at), score: 0.78 POMZP3POM (POM121 homolog, rat) and ZP3 fusion (204148_s_at), score: 0.7 PRKCDprotein kinase C, delta (202545_at), score: 0.71 PRMT3protein arginine methyltransferase 3 (213320_at), score: -0.82 PTPN9protein tyrosine phosphatase, non-receptor type 9 (202958_at), score: 0.65 RAB1ARAB1A, member RAS oncogene family (213440_at), score: -0.7 RAB33BRAB33B, member RAS oncogene family (221014_s_at), score: -0.65 RAD54L2RAD54-like 2 (S. cerevisiae) (213205_s_at), score: 0.69 RBM15BRNA binding motif protein 15B (202689_at), score: 0.69 REREarginine-glutamic acid dipeptide (RE) repeats (200940_s_at), score: 0.75 RNF11ring finger protein 11 (208924_at), score: -0.77 RNF220ring finger protein 220 (219988_s_at), score: 0.91 RPL18AP6ribosomal protein L18a pseudogene 6 (216383_at), score: -0.92 RPRD2regulation of nuclear pre-mRNA domain containing 2 (212553_at), score: 0.65 RPS17P5ribosomal protein S17 pseudogene 5 (216348_at), score: -0.86 RPS25ribosomal protein S25 (200091_s_at), score: -0.66 SAP30LSAP30-like (219129_s_at), score: 0.67 SBNO2strawberry notch homolog 2 (Drosophila) (204166_at), score: 0.74 SCAMP4secretory carrier membrane protein 4 (213244_at), score: 1 SDC1syndecan 1 (201287_s_at), score: 0.63 SF1splicing factor 1 (208313_s_at), score: 0.89 SF3A2splicing factor 3a, subunit 2, 66kDa (37462_i_at), score: 0.7 SIRT6sirtuin (silent mating type information regulation 2 homolog) 6 (S. cerevisiae) (219613_s_at), score: 0.63 SLC33A1solute carrier family 33 (acetyl-CoA transporter), member 1 (203165_s_at), score: -0.76 SLC43A3solute carrier family 43, member 3 (213113_s_at), score: 0.76 SMG6Smg-6 homolog, nonsense mediated mRNA decay factor (C. elegans) (214940_s_at), score: 0.69 SMG7Smg-7 homolog, nonsense mediated mRNA decay factor (C. elegans) (201794_s_at), score: 0.68 SNAP23synaptosomal-associated protein, 23kDa (214544_s_at), score: -0.67 SNAP25synaptosomal-associated protein, 25kDa (202508_s_at), score: 0.63 SOLHsmall optic lobes homolog (Drosophila) (204275_at), score: 0.85 SPG7spastic paraplegia 7 (pure and complicated autosomal recessive) (202104_s_at), score: 0.66 STIM1stromal interaction molecule 1 (202764_at), score: 0.69 STRN4striatin, calmodulin binding protein 4 (217903_at), score: 0.74 SULT1B1sulfotransferase family, cytosolic, 1B, member 1 (207601_at), score: -0.74 TAPBPTAP binding protein (tapasin) (208829_at), score: 0.73 TBCDtubulin folding cofactor D (211052_s_at), score: 0.7 TIAF1TGFB1-induced anti-apoptotic factor 1 (202039_at), score: 0.72 TICAM2toll-like receptor adaptor molecule 2 (214658_at), score: -0.85 TLE4transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila) (204872_at), score: -0.65 TMED7transmembrane emp24 protein transport domain containing 7 (209404_s_at), score: -0.7 TMEM123transmembrane protein 123 (211967_at), score: -0.65 TRIM21tripartite motif-containing 21 (204804_at), score: 0.64 TXLNAtaxilin alpha (212300_at), score: 0.8 UBTD1ubiquitin domain containing 1 (219172_at), score: 0.72 UGDHUDP-glucose dehydrogenase (203343_at), score: -0.65 USF2upstream transcription factor 2, c-fos interacting (202152_x_at), score: 0.66 VEGFBvascular endothelial growth factor B (203683_s_at), score: 0.81 VPS37Bvacuolar protein sorting 37 homolog B (S. cerevisiae) (221704_s_at), score: 0.78 WDR6WD repeat domain 6 (217734_s_at), score: 0.81 WDR62WD repeat domain 62 (215218_s_at), score: 0.69 XAB2XPA binding protein 2 (218110_at), score: 0.72 YES1v-yes-1 Yamaguchi sarcoma viral oncogene homolog 1 (202932_at), score: 0.71 YTHDC2YTH domain containing 2 (213077_at), score: -0.67 ZFP36L1zinc finger protein 36, C3H type-like 1 (211965_at), score: 0.68 ZFYVE26zinc finger, FYVE domain containing 26 (37943_at), score: -0.66 ZNF318zinc finger protein 318 (203521_s_at), score: 0.72 ZNF536zinc finger protein 536 (206403_at), score: 0.65
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-TABM-263-raw-cel-1515485771.cel | 7 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486371.cel | 37 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485891.cel | 13 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485811.cel | 9 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485751.cel | 6 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486091.cel | 23 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486431.cel | 40 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485671.cel | 2 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486031.cel | 20 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486071.cel | 22 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485711.cel | 4 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486411.cel | 39 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-GEOD-3860-raw-cel-1561690344.cel | 10 | 5 | HGPS | hgu133a | none | GM00038C |
| E-TABM-263-raw-cel-1515485871.cel | 12 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485691.cel | 3 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-GEOD-3860-raw-cel-1561690304.cel | 8 | 5 | HGPS | hgu133a | none | GMO8398C |
| E-TABM-263-raw-cel-1515486211.cel | 29 | 6 | Cycle | hgu133a2 | none | Cycle 1 |