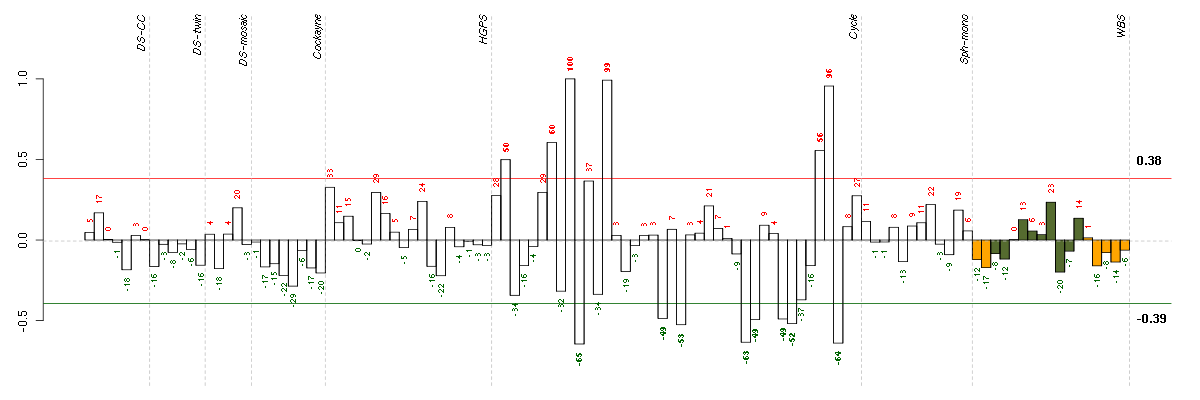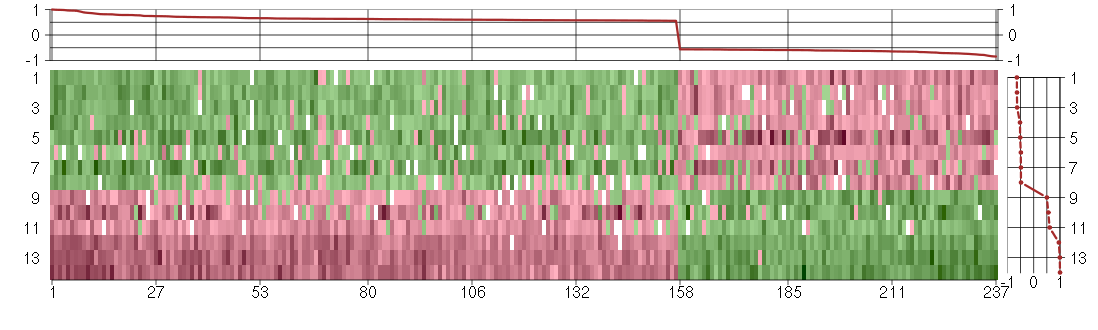

Under-expression is coded with green,
over-expression with red color.

digestion
The whole of the physical, chemical, and biochemical processes carried out by multicellular organisms to break down ingested nutrients into components that may be easily absorbed and directed into metabolism.
biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
multicellular organismal process
Any biological process, occurring at the level of a multicellular organism, pertinent to its function.
all
This term is the most general term possible

plasma membrane
The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins.
membrane
Double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
cell
The basic structural and functional unit of all organisms. Includes the plasma membrane and any external encapsulating structures such as the cell wall and cell envelope.
intrinsic to membrane
Located in a membrane such that some covalently attached portion of the gene product, for example part of a peptide sequence or some other covalently attached moiety such as a GPI anchor, spans or is embedded in one or both leaflets of the membrane.
membrane part
Any constituent part of a membrane, a double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
all
This term is the most general term possible
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
membrane part
Any constituent part of a membrane, a double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.

purinergic nucleotide receptor activity
Combining with a purine nucleotide to initiate a change in cell activity.
molecular_function
Elemental activities, such as catalysis or binding, describing the actions of a gene product at the molecular level. A given gene product may exhibit one or more molecular functions.
signal transducer activity
Mediates the transfer of a signal from the outside to the inside of a cell by means other than the introduction of the signal molecule itself into the cell.
receptor activity
Combining with an extracellular or intracellular messenger to initiate a change in cell activity.
transmembrane receptor activity
Combining with an extracellular or intracellular messenger to initiate a change in cell activity, and spanning to the membrane of either the cell or an organelle.
nucleotide receptor activity
Combining with a nucleotide to initiate a change in cell activity. A nucleotide is a compound that consists of a nucleoside esterified with a phosphate molecule.
molecular transducer activity
The molecular function that accepts an input of one form and creates an output of a different form.
all
This term is the most general term possible
| Id | Pvalue | ExpCount | Count | Size | Term |
|---|---|---|---|---|---|
| 04080 | 6.132e-04 | 2.466 | 12 | 83 | Neuroactive ligand-receptor interaction |
ABCE1ATP-binding cassette, sub-family E (OABP), member 1 (201873_s_at), score: 0.56 ACSL1acyl-CoA synthetase long-chain family member 1 (201963_at), score: 0.56 ACSL4acyl-CoA synthetase long-chain family member 4 (202422_s_at), score: 0.74 ACSS3acyl-CoA synthetase short-chain family member 3 (219616_at), score: -0.63 ACTR3BARP3 actin-related protein 3 homolog B (yeast) (218868_at), score: 0.66 ADAM19ADAM metallopeptidase domain 19 (meltrin beta) (209765_at), score: 0.58 ADARadenosine deaminase, RNA-specific (201786_s_at), score: -0.63 ADORA2Badenosine A2b receptor (205891_at), score: -0.84 AHI1Abelson helper integration site 1 (221569_at), score: 0.92 AIM1absent in melanoma 1 (212543_at), score: -0.72 AJAP1adherens junctions associated protein 1 (206460_at), score: 0.69 ALDH3A2aldehyde dehydrogenase 3 family, member A2 (202053_s_at), score: -0.62 ANXA10annexin A10 (210143_at), score: 0.98 APAF1apoptotic peptidase activating factor 1 (204859_s_at), score: 0.68 APOBEC3Fapolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 3F (214995_s_at), score: -0.64 APOL6apolipoprotein L, 6 (219716_at), score: -0.65 ARHGEF2rho/rac guanine nucleotide exchange factor (GEF) 2 (209435_s_at), score: -0.61 ARL4CADP-ribosylation factor-like 4C (202207_at), score: -0.65 ARMC9armadillo repeat containing 9 (219637_at), score: -0.65 ARNT2aryl-hydrocarbon receptor nuclear translocator 2 (202986_at), score: -0.6 ATP2C1ATPase, Ca++ transporting, type 2C, member 1 (211137_s_at), score: 0.59 ATP6V1B2ATPase, H+ transporting, lysosomal 56/58kDa, V1 subunit B2 (201089_at), score: -0.59 BMP6bone morphogenetic protein 6 (206176_at), score: 0.87 BTBD3BTB (POZ) domain containing 3 (202946_s_at), score: 0.56 C14orf45chromosome 14 open reading frame 45 (220173_at), score: 0.63 C1orf9chromosome 1 open reading frame 9 (203429_s_at), score: 0.66 C2CD2C2 calcium-dependent domain containing 2 (212875_s_at), score: 0.58 C2CD2LC2CD2-like (204757_s_at), score: 0.6 C4orf43chromosome 4 open reading frame 43 (218513_at), score: 0.69 CALB2calbindin 2 (205428_s_at), score: 0.62 CCDC28Acoiled-coil domain containing 28A (209479_at), score: -0.69 CCNG1cyclin G1 (208796_s_at), score: -0.56 CCNG2cyclin G2 (211559_s_at), score: -0.59 CCNT2cyclin T2 (204645_at), score: 0.63 CD55CD55 molecule, decay accelerating factor for complement (Cromer blood group) (201925_s_at), score: 0.75 CDC2L6cell division cycle 2-like 6 (CDK8-like) (212899_at), score: -0.72 CDCP1CUB domain containing protein 1 (218451_at), score: 0.65 CDH6cadherin 6, type 2, K-cadherin (fetal kidney) (205532_s_at), score: 0.6 CEP68centrosomal protein 68kDa (212677_s_at), score: -0.62 CEP70centrosomal protein 70kDa (219036_at), score: -0.61 CHUKconserved helix-loop-helix ubiquitous kinase (209666_s_at), score: 0.66 CMPK1cytidine monophosphate (UMP-CMP) kinase 1, cytosolic (217870_s_at), score: 0.6 COL5A3collagen, type V, alpha 3 (52255_s_at), score: 0.56 CRBNcereblon (218142_s_at), score: -0.58 CRTC3CREB regulated transcription coactivator 3 (218648_at), score: -0.76 CSGALNACT2chondroitin sulfate N-acetylgalactosaminyltransferase 2 (222235_s_at), score: 0.73 CST6cystatin E/M (206595_at), score: 0.59 CXCL6chemokine (C-X-C motif) ligand 6 (granulocyte chemotactic protein 2) (206336_at), score: 0.71 DCP2DCP2 decapping enzyme homolog (S. cerevisiae) (212919_at), score: 0.64 DENND5BDENN/MADD domain containing 5B (215058_at), score: 0.63 DEPDC6DEP domain containing 6 (218858_at), score: -0.78 DIRAS3DIRAS family, GTP-binding RAS-like 3 (215506_s_at), score: 0.78 DIS3DIS3 mitotic control homolog (S. cerevisiae) (218362_s_at), score: 0.58 DNAJB9DnaJ (Hsp40) homolog, subfamily B, member 9 (202843_at), score: 0.95 DOCK10dedicator of cytokinesis 10 (219279_at), score: 0.6 DUSP4dual specificity phosphatase 4 (204014_at), score: 0.81 DZIP3DAZ interacting protein 3, zinc finger (213186_at), score: -0.65 EAF2ELL associated factor 2 (219551_at), score: 0.64 EDEM1ER degradation enhancer, mannosidase alpha-like 1 (203279_at), score: 0.57 EGFepidermal growth factor (beta-urogastrone) (206254_at), score: 0.62 EIF2S1eukaryotic translation initiation factor 2, subunit 1 alpha, 35kDa (201143_s_at), score: 0.62 EIF4Eeukaryotic translation initiation factor 4E (201436_at), score: 0.69 ELK3ELK3, ETS-domain protein (SRF accessory protein 2) (221773_at), score: 0.62 EML1echinoderm microtubule associated protein like 1 (204797_s_at), score: 0.58 ESM1endothelial cell-specific molecule 1 (208394_x_at), score: 0.88 ESRRAestrogen-related receptor alpha (1487_at), score: 0.69 EVI2Becotropic viral integration site 2B (211742_s_at), score: -0.59 FAM18Bfamily with sequence similarity 18, member B (218446_s_at), score: 0.58 FBXO31F-box protein 31 (219785_s_at), score: -0.57 FERMT1fermitin family homolog 1 (Drosophila) (218796_at), score: 0.81 FGF1fibroblast growth factor 1 (acidic) (205117_at), score: 0.57 FGF7fibroblast growth factor 7 (keratinocyte growth factor) (205782_at), score: 0.58 FICDFIC domain containing (219910_at), score: 0.67 FNBP1formin binding protein 1 (212288_at), score: -0.68 GABBR2gamma-aminobutyric acid (GABA) B receptor, 2 (209990_s_at), score: 0.6 GALgalanin prepropeptide (214240_at), score: 0.58 GALK2galactokinase 2 (205219_s_at), score: 0.57 GFOD1glucose-fructose oxidoreductase domain containing 1 (219821_s_at), score: 0.7 GHRgrowth hormone receptor (205498_at), score: 0.63 GKglycerol kinase (207387_s_at), score: 0.99 GK3Pglycerol kinase 3 pseudogene (215966_x_at), score: 0.96 GMCL1germ cell-less homolog 1 (Drosophila) (218458_at), score: 0.74 GOLT1Bgolgi transport 1 homolog B (S. cerevisiae) (218193_s_at), score: 0.58 GPM6Bglycoprotein M6B (209170_s_at), score: 0.75 GPR177G protein-coupled receptor 177 (221958_s_at), score: 0.78 GPR183G protein-coupled receptor 183 (205419_at), score: 0.57 GRAMD1BGRAM domain containing 1B (212906_at), score: 0.57 GRB14growth factor receptor-bound protein 14 (206204_at), score: 0.71 GRSF1G-rich RNA sequence binding factor 1 (201501_s_at), score: 0.57 GTF2A1Lgeneral transcription factor IIA, 1-like (213413_at), score: -0.81 H1FXH1 histone family, member X (204805_s_at), score: -0.57 HDGFhepatoma-derived growth factor (high-mobility group protein 1-like) (200896_x_at), score: -0.62 HISPPD1histidine acid phosphatase domain containing 1 (203253_s_at), score: 0.61 HSD17B6hydroxysteroid (17-beta) dehydrogenase 6 homolog (mouse) (37512_at), score: 0.63 HTR2A5-hydroxytryptamine (serotonin) receptor 2A (207135_at), score: 0.77 IL11interleukin 11 (206924_at), score: 0.6 IL13RA2interleukin 13 receptor, alpha 2 (206172_at), score: 0.72 IL33interleukin 33 (209821_at), score: 0.99 IRGQimmunity-related GTPase family, Q (64488_at), score: 0.57 ITGAVintegrin, alpha V (vitronectin receptor, alpha polypeptide, antigen CD51) (202351_at), score: 0.58 KAL1Kallmann syndrome 1 sequence (205206_at), score: 1 KCNG1potassium voltage-gated channel, subfamily G, member 1 (214595_at), score: 0.62 KIAA0562KIAA0562 (204075_s_at), score: 0.58 KIAA1305KIAA1305 (220911_s_at), score: -0.62 KIAA1462KIAA1462 (213316_at), score: -0.73 KLHL21kelch-like 21 (Drosophila) (203068_at), score: 0.63 KLHL23kelch-like 23 (Drosophila) (213610_s_at), score: 0.58 LARP4La ribonucleoprotein domain family, member 4 (214155_s_at), score: 0.63 LDLRlow density lipoprotein receptor (202068_s_at), score: -0.57 LRP8low density lipoprotein receptor-related protein 8, apolipoprotein e receptor (205282_at), score: 0.74 LYPD1LY6/PLAUR domain containing 1 (212909_at), score: 0.61 MAN1C1mannosidase, alpha, class 1C, member 1 (218918_at), score: -0.59 MARCH3membrane-associated ring finger (C3HC4) 3 (213256_at), score: 0.57 MC4Rmelanocortin 4 receptor (221467_at), score: 0.57 MFAP3Lmicrofibrillar-associated protein 3-like (205442_at), score: 0.63 MIS12MIS12, MIND kinetochore complex component, homolog (yeast) (221559_s_at), score: 0.6 MKNK2MAP kinase interacting serine/threonine kinase 2 (218205_s_at), score: -0.57 MMP10matrix metallopeptidase 10 (stromelysin 2) (205680_at), score: 0.8 MMP16matrix metallopeptidase 16 (membrane-inserted) (207012_at), score: 0.71 MREGmelanoregulin (219648_at), score: 0.62 MSTO1misato homolog 1 (Drosophila) (218296_x_at), score: 0.6 MTAPmethylthioadenosine phosphorylase (211363_s_at), score: 0.57 MTMR9myotubularin related protein 9 (204837_at), score: 0.57 NAGPAN-acetylglucosamine-1-phosphodiester alpha-N-acetylglucosaminidase (205090_s_at), score: -0.57 NAT13N-acetyltransferase 13 (GCN5-related) (217745_s_at), score: 0.6 NCALDneurocalcin delta (211685_s_at), score: 0.81 NDRG4NDRG family member 4 (209159_s_at), score: -0.61 NDUFA2NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 2, 8kDa (213550_s_at), score: -0.62 NEFLneurofilament, light polypeptide (221805_at), score: 0.79 NEFMneurofilament, medium polypeptide (205113_at), score: 0.96 NMIN-myc (and STAT) interactor (203964_at), score: -0.58 NOX4NADPH oxidase 4 (219773_at), score: 0.65 NPIPnuclear pore complex interacting protein (204538_x_at), score: -0.61 NPTX1neuronal pentraxin I (204684_at), score: 0.84 NR5A2nuclear receptor subfamily 5, group A, member 2 (208343_s_at), score: 0.6 NRXN3neurexin 3 (205795_at), score: 0.61 NTMneurotrimin (222020_s_at), score: 0.63 OBFC2Aoligonucleotide/oligosaccharide-binding fold containing 2A (219334_s_at), score: 0.58 OPCMLopioid binding protein/cell adhesion molecule-like (214111_at), score: 0.77 ORAI3ORAI calcium release-activated calcium modulator 3 (221864_at), score: -0.59 OSTM1osteopetrosis associated transmembrane protein 1 (218196_at), score: 0.64 P2RX5purinergic receptor P2X, ligand-gated ion channel, 5 (210448_s_at), score: 0.66 P2RY5purinergic receptor P2Y, G-protein coupled, 5 (218589_at), score: -0.65 PAQR3progestin and adipoQ receptor family member III (213372_at), score: 0.66 PCDH9protocadherin 9 (219737_s_at), score: 0.86 PCTK2PCTAIRE protein kinase 2 (221918_at), score: 0.62 PCYT2phosphate cytidylyltransferase 2, ethanolamine (209577_at), score: -0.59 PDCD1LG2programmed cell death 1 ligand 2 (220049_s_at), score: 0.63 PDE5Aphosphodiesterase 5A, cGMP-specific (206757_at), score: -0.63 PECRperoxisomal trans-2-enoyl-CoA reductase (221142_s_at), score: -0.62 PET112LPET112-like (yeast) (204300_at), score: -0.59 PHKA1phosphorylase kinase, alpha 1 (muscle) (205450_at), score: 0.63 PID1phosphotyrosine interaction domain containing 1 (219093_at), score: 0.56 PIGHphosphatidylinositol glycan anchor biosynthesis, class H (209625_at), score: 0.61 PIGLphosphatidylinositol glycan anchor biosynthesis, class L (213889_at), score: -0.65 PIONpigeon homolog (Drosophila) (222150_s_at), score: 0.67 PITX1paired-like homeodomain 1 (208502_s_at), score: -0.58 PLA2G4Aphospholipase A2, group IVA (cytosolic, calcium-dependent) (210145_at), score: 0.58 PMAIP1phorbol-12-myristate-13-acetate-induced protein 1 (204286_s_at), score: 0.6 PMP22peripheral myelin protein 22 (210139_s_at), score: -0.57 PPARGperoxisome proliferator-activated receptor gamma (208510_s_at), score: -0.61 PRMT3protein arginine methyltransferase 3 (213320_at), score: 0.6 PRSS1protease, serine, 1 (trypsin 1) (216470_x_at), score: 0.64 PRSS2protease, serine, 2 (trypsin 2) (205402_x_at), score: 0.68 PRSS3protease, serine, 3 (207463_x_at), score: 0.74 PTGS1prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) (215813_s_at), score: 0.64 PTGS2prostaglandin-endoperoxide synthase 2 (prostaglandin G/H synthase and cyclooxygenase) (204748_at), score: 0.62 PTHLHparathyroid hormone-like hormone (211756_at), score: 0.7 PTTG3pituitary tumor-transforming 3 (208511_at), score: -0.57 PVRL3poliovirus receptor-related 3 (213325_at), score: 0.59 RAB6CRAB6C, member RAS oncogene family (210406_s_at), score: 0.57 RAB7ARAB7A, member RAS oncogene family (211960_s_at), score: 0.64 RAB8BRAB8B, member RAS oncogene family (219210_s_at), score: -0.56 RAGErenal tumor antigen (205130_at), score: 0.56 RBM3RNA binding motif (RNP1, RRM) protein 3 (208319_s_at), score: 0.6 REREarginine-glutamic acid dipeptide (RE) repeats (200940_s_at), score: -0.59 RFX5regulatory factor X, 5 (influences HLA class II expression) (202963_at), score: -0.71 RNF44ring finger protein 44 (203286_at), score: -0.74 RP2retinitis pigmentosa 2 (X-linked recessive) (205191_at), score: 0.64 RPL21P68ribosomal protein L21 pseudogene 68 (217340_at), score: 0.58 RPL23ribosomal protein L23 (214744_s_at), score: 0.66 RPLP2P1ribosomal protein, large P2, pseudogene 1 (216490_x_at), score: 0.61 RPS11ribosomal protein S11 (213350_at), score: -0.56 S1PR1sphingosine-1-phosphate receptor 1 (204642_at), score: 0.63 SAV1salvador homolog 1 (Drosophila) (218276_s_at), score: -0.58 SERPINB2serpin peptidase inhibitor, clade B (ovalbumin), member 2 (204614_at), score: 0.69 SERPINB9serpin peptidase inhibitor, clade B (ovalbumin), member 9 (209723_at), score: -0.65 SESN1sestrin 1 (218346_s_at), score: -0.58 SIGLEC6sialic acid binding Ig-like lectin 6 (210796_x_at), score: 0.56 SLC2A6solute carrier family 2 (facilitated glucose transporter), member 6 (220091_at), score: -0.57 SLC33A1solute carrier family 33 (acetyl-CoA transporter), member 1 (203165_s_at), score: 0.63 SLC38A4solute carrier family 38, member 4 (220786_s_at), score: -0.71 SLC4A4solute carrier family 4, sodium bicarbonate cotransporter, member 4 (203908_at), score: 0.6 SLC4A7solute carrier family 4, sodium bicarbonate cotransporter, member 7 (209884_s_at), score: 0.82 SLMO2slowmo homolog 2 (Drosophila) (217851_s_at), score: 0.71 SMAD3SMAD family member 3 (218284_at), score: -0.7 SNNstannin (218032_at), score: -0.59 SPA17sperm autoantigenic protein 17 (205406_s_at), score: -0.59 SS18L1synovial sarcoma translocation gene on chromosome 18-like 1 (213140_s_at), score: 0.56 SSTR1somatostatin receptor 1 (208482_at), score: 0.7 ST5suppression of tumorigenicity 5 (202440_s_at), score: -0.67 STARD5StAR-related lipid transfer (START) domain containing 5 (213820_s_at), score: -0.59 SYNJ1synaptojanin 1 (212990_at), score: 0.61 TCEB1transcription elongation factor B (SIII), polypeptide 1 (15kDa, elongin C) (202823_at), score: 0.57 TENC1tensin like C1 domain containing phosphatase (tensin 2) (212494_at), score: -0.61 TFDP2transcription factor Dp-2 (E2F dimerization partner 2) (203588_s_at), score: -0.85 TGDSTDP-glucose 4,6-dehydratase (208249_s_at), score: 0.7 TGFAtransforming growth factor, alpha (205016_at), score: 0.62 THYN1thymocyte nuclear protein 1 (218491_s_at), score: -0.59 TICAM2toll-like receptor adaptor molecule 2 (214658_at), score: 0.64 TLR3toll-like receptor 3 (206271_at), score: -0.61 TM4SF1transmembrane 4 L six family member 1 (209386_at), score: 0.58 TMEFF1transmembrane protein with EGF-like and two follistatin-like domains 1 (205122_at), score: 0.58 TMEM2transmembrane protein 2 (218113_at), score: 0.78 TMSB15Athymosin beta 15a (205347_s_at), score: -0.63 TMSB15Bthymosin beta 15B (214051_at), score: -0.58 TNS3tensin 3 (217853_at), score: -0.62 TOMM20translocase of outer mitochondrial membrane 20 homolog (yeast) (200662_s_at), score: 0.65 TP53tumor protein p53 (201746_at), score: -0.78 TRIM21tripartite motif-containing 21 (204804_at), score: -0.58 TRPC4transient receptor potential cation channel, subfamily C, member 4 (220817_at), score: 0.6 TSPAN13tetraspanin 13 (217979_at), score: 0.63 UBXN1UBX domain protein 1 (201871_s_at), score: -0.56 UCP2uncoupling protein 2 (mitochondrial, proton carrier) (208998_at), score: -0.72 UFM1ubiquitin-fold modifier 1 (218050_at), score: 0.65 UGDHUDP-glucose dehydrogenase (203343_at), score: 0.59 UHRF1BP1LUHRF1 binding protein 1-like (213118_at), score: 0.7 ULBP2UL16 binding protein 2 (221291_at), score: 0.59 URB1URB1 ribosome biogenesis 1 homolog (S. cerevisiae) (212996_s_at), score: 0.61 VWA5Avon Willebrand factor A domain containing 5A (210102_at), score: -0.75 WDR6WD repeat domain 6 (217734_s_at), score: -0.67 XYLT1xylosyltransferase I (213725_x_at), score: 0.6 YDD19YDD19 protein (37079_at), score: 0.63 ZDHHC7zinc finger, DHHC-type containing 7 (218606_at), score: -0.69 ZFYVE16zinc finger, FYVE domain containing 16 (203651_at), score: 0.58 ZNF395zinc finger protein 395 (218149_s_at), score: -0.56 ZNF706zinc finger protein 706 (218059_at), score: -0.56
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-TABM-263-raw-cel-1515485831.cel | 10 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486391.cel | 38 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486191.cel | 28 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486051.cel | 21 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486291.cel | 33 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486211.cel | 29 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486271.cel | 32 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486011.cel | 19 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485671.cel | 2 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486351.cel | 36 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485771.cel | 7 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486371.cel | 37 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485891.cel | 13 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485811.cel | 9 | 6 | Cycle | hgu133a2 | none | Cycle 1 |