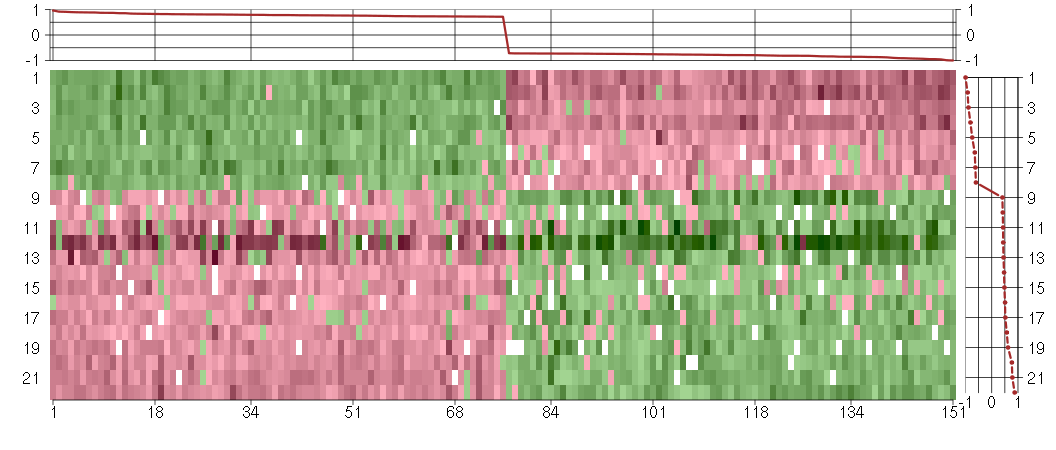

Under-expression is coded with green,
over-expression with red color.

protein import into nucleus
The directed movement of a protein from the cytoplasm to the nucleus.
signal transduction
The cascade of processes by which a signal interacts with a receptor, causing a change in the level or activity of a second messenger or other downstream target, and ultimately effecting a change in the functioning of the cell.
protein localization
Any process by which a protein is transported to, or maintained in, a specific location.
protein targeting
The process of targeting specific proteins to particular membrane-bounded subcellular organelles. Usually requires an organelle specific protein sequence motif.
transport
The directed movement of substances (such as macromolecules, small molecules, ions) into, out of, within or between cells, or within a multicellular organism.
intracellular protein transport
The directed movement of proteins in a cell, including the movement of proteins between specific compartments or structures within a cell, such as organelles of a eukaryotic cell.
nucleocytoplasmic transport
The directed movement of molecules between the nucleus and the cytoplasm.
cell communication
Any process that mediates interactions between a cell and its surroundings. Encompasses interactions such as signaling or attachment between one cell and another cell, between a cell and an extracellular matrix, or between a cell and any other aspect of its environment.
intracellular signaling cascade
A series of reactions within the cell that occur as a result of a single trigger reaction or compound.
protein kinase cascade
A series of reactions, mediated by protein kinases, which occurs as a result of a single trigger reaction or compound.
I-kappaB kinase/NF-kappaB cascade
A series of reactions initiated by the activation of the transcription factor NF-kappaB. NF-kappaB is sequestered by the inhibitor I-kappaB, and is released when I-kappaB is phosphorylated by activated I-kappaB kinase.
biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
cellular process
Any process that is carried out at the cellular level, but not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
protein transport
The directed movement of proteins into, out of, within or between cells.
protein import
The directed movement of proteins into a cell or organelle.
regulation of intracellular transport
Any process that modulates the frequency, rate or extent of the directed movement of substances within cells.
negative regulation of intracellular transport
Any process that stops, prevents, or reduces the frequency, rate or extent of the directed movement of substances within cells.
regulation of localization
Any process that modulates the frequency, rate or extent of any process by which a cell, a substance, or a cellular entity is transported to, or maintained in, a specific location.
regulation of protein localization
Any process that modulates the frequency, rate or extent of any process by which a protein is transported to, or maintained in, a specific location.
macromolecule localization
Any process by which a macromolecule is transported to, or maintained in, a specific location.
regulation of intracellular protein transport
Any process that modulates the frequency, rate or extent of the directed movement of proteins within cells.
cellular protein localization
Any process by which a protein is transported to, and/or maintained in, a specific location within or at the surface of a cell.
regulation of protein import into nucleus
Any process that modulates the frequency, rate or extent of movement of proteins from the cytoplasm to the nucleus.
negative regulation of protein import into nucleus
Any process that stops, prevents or reduces the frequency, rate or extent of the movement of proteins from the cytoplasm into the nucleus.
regulation of transcription factor import into nucleus
Any process that modulates the frequency, rate or extent of the movement of a transcription factor from the cytoplasm to the nucleus.
transcription factor import into nucleus
The directed movement of a transcription factor from the cytoplasm to the nucleus.
negative regulation of transcription factor import into nucleus
Any process that stops, prevents or reduces the frequency, rate or extent of the movement of a transcription factor from the cytoplasm to the nucleus.
establishment of protein localization
The directed movement of a protein to a specific location.
regulation of nucleocytoplasmic transport
Any process that modulates the frequency, rate or extent of the directed movement of substances between the nucleus and the cytoplasm.
negative regulation of nucleocytoplasmic transport
Any process that stops, prevents or reduces the frequency, rate or extent of the directed movement of substances between the cytoplasm and the nucleus.
intracellular transport
The directed movement of substances within a cell.
negative regulation of biological process
Any process that stops, prevents or reduces the frequency, rate or extent of a biological process. Biological processes are regulated by many means; examples include the control of gene expression, protein modification or interaction with a protein or substrate molecule.
regulation of biological process
Any process that modulates the frequency, rate or extent of a biological process. Biological processes are regulated by many means; examples include the control of gene expression, protein modification or interaction with a protein or substrate molecule.
regulation of cellular process
Any process that modulates the frequency, rate or extent of a cellular process, any of those that are carried out at the cellular level, but are not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
regulation of transport
Any process that modulates the frequency, rate or extent of the directed movement of substances (such as macromolecules, small molecules, ions) into, out of, within or between cells.
negative regulation of transport
Any process that stops, prevents or reduces the frequency, rate or extent of the directed movement of substances (such as macromolecules, small molecules, ions) into, out of, within or between cells.
nuclear transport
The directed movement of substances into, out of, or within the nucleus.
nuclear import
The directed movement of substances into the nucleus.
localization
Any process by which a cell, a substance, or a cellular entity, such as a protein complex or organelle, is transported to, and/or maintained in a specific location.
regulation of protein transport
Any process that modulates the frequency, rate or extent of the directed movement of a protein into, out of, within or between cells.
negative regulation of protein transport
Any process that stops, prevents or reduces the frequency, rate or extent of the directed movement of a protein into, out of, within or between cells.
establishment of localization
The directed movement of a cell, substance or cellular entity, such as a protein complex or organelle, to a specific location.
cellular localization
Any process by which a substance or cellular entity, such as a protein complex or organelle, is transported to, and/or maintained in, a specific location within or in the membrane of a cell.
establishment of localization in cell
The directed movement of a substance or cellular entity, such as a protein complex or organelle, to a specific location within, or in the membrane of, a cell.
regulation of cellular localization
Any process that modulates the frequency, rate or extent of a process by which a cell, a substance, or a cellular entity is transported to, or maintained in a specific location within or in the membrane of a cell.
biological regulation
Any process that modulates the frequency, rate or extent of any biological process, quality or function.
regulation of establishment of protein localization
Any process that modulates the frequency, rate or extent of the directed movement of a protein to a specific location.
all
This term is the most general term possible
negative regulation of biological process
Any process that stops, prevents or reduces the frequency, rate or extent of a biological process. Biological processes are regulated by many means; examples include the control of gene expression, protein modification or interaction with a protein or substrate molecule.
regulation of cellular process
Any process that modulates the frequency, rate or extent of a cellular process, any of those that are carried out at the cellular level, but are not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
regulation of localization
Any process that modulates the frequency, rate or extent of any process by which a cell, a substance, or a cellular entity is transported to, or maintained in, a specific location.
establishment of localization
The directed movement of a cell, substance or cellular entity, such as a protein complex or organelle, to a specific location.
cellular localization
Any process by which a substance or cellular entity, such as a protein complex or organelle, is transported to, and/or maintained in, a specific location within or in the membrane of a cell.
regulation of biological process
Any process that modulates the frequency, rate or extent of a biological process. Biological processes are regulated by many means; examples include the control of gene expression, protein modification or interaction with a protein or substrate molecule.
signal transduction
The cascade of processes by which a signal interacts with a receptor, causing a change in the level or activity of a second messenger or other downstream target, and ultimately effecting a change in the functioning of the cell.
establishment of localization in cell
The directed movement of a substance or cellular entity, such as a protein complex or organelle, to a specific location within, or in the membrane of, a cell.
regulation of cellular localization
Any process that modulates the frequency, rate or extent of a process by which a cell, a substance, or a cellular entity is transported to, or maintained in a specific location within or in the membrane of a cell.
regulation of cellular localization
Any process that modulates the frequency, rate or extent of a process by which a cell, a substance, or a cellular entity is transported to, or maintained in a specific location within or in the membrane of a cell.
regulation of transport
Any process that modulates the frequency, rate or extent of the directed movement of substances (such as macromolecules, small molecules, ions) into, out of, within or between cells.
negative regulation of transport
Any process that stops, prevents or reduces the frequency, rate or extent of the directed movement of substances (such as macromolecules, small molecules, ions) into, out of, within or between cells.
protein transport
The directed movement of proteins into, out of, within or between cells.
intracellular transport
The directed movement of substances within a cell.
regulation of establishment of protein localization
Any process that modulates the frequency, rate or extent of the directed movement of a protein to a specific location.
regulation of intracellular transport
Any process that modulates the frequency, rate or extent of the directed movement of substances within cells.
negative regulation of transport
Any process that stops, prevents or reduces the frequency, rate or extent of the directed movement of substances (such as macromolecules, small molecules, ions) into, out of, within or between cells.
regulation of protein localization
Any process that modulates the frequency, rate or extent of any process by which a protein is transported to, or maintained in, a specific location.
cellular protein localization
Any process by which a protein is transported to, and/or maintained in, a specific location within or at the surface of a cell.
establishment of protein localization
The directed movement of a protein to a specific location.
intracellular protein transport
The directed movement of proteins in a cell, including the movement of proteins between specific compartments or structures within a cell, such as organelles of a eukaryotic cell.
regulation of protein transport
Any process that modulates the frequency, rate or extent of the directed movement of a protein into, out of, within or between cells.
negative regulation of protein transport
Any process that stops, prevents or reduces the frequency, rate or extent of the directed movement of a protein into, out of, within or between cells.
intracellular protein transport
The directed movement of proteins in a cell, including the movement of proteins between specific compartments or structures within a cell, such as organelles of a eukaryotic cell.
regulation of intracellular transport
Any process that modulates the frequency, rate or extent of the directed movement of substances within cells.
negative regulation of intracellular transport
Any process that stops, prevents, or reduces the frequency, rate or extent of the directed movement of substances within cells.
regulation of protein transport
Any process that modulates the frequency, rate or extent of the directed movement of a protein into, out of, within or between cells.
negative regulation of intracellular transport
Any process that stops, prevents, or reduces the frequency, rate or extent of the directed movement of substances within cells.
regulation of intracellular protein transport
Any process that modulates the frequency, rate or extent of the directed movement of proteins within cells.
negative regulation of protein import into nucleus
Any process that stops, prevents or reduces the frequency, rate or extent of the movement of proteins from the cytoplasm into the nucleus.
regulation of intracellular protein transport
Any process that modulates the frequency, rate or extent of the directed movement of proteins within cells.
negative regulation of protein transport
Any process that stops, prevents or reduces the frequency, rate or extent of the directed movement of a protein into, out of, within or between cells.
regulation of protein import into nucleus
Any process that modulates the frequency, rate or extent of movement of proteins from the cytoplasm to the nucleus.
negative regulation of nucleocytoplasmic transport
Any process that stops, prevents or reduces the frequency, rate or extent of the directed movement of substances between the cytoplasm and the nucleus.
protein import into nucleus
The directed movement of a protein from the cytoplasm to the nucleus.
regulation of protein import into nucleus
Any process that modulates the frequency, rate or extent of movement of proteins from the cytoplasm to the nucleus.
negative regulation of protein import into nucleus
Any process that stops, prevents or reduces the frequency, rate or extent of the movement of proteins from the cytoplasm into the nucleus.
regulation of nucleocytoplasmic transport
Any process that modulates the frequency, rate or extent of the directed movement of substances between the nucleus and the cytoplasm.
negative regulation of nucleocytoplasmic transport
Any process that stops, prevents or reduces the frequency, rate or extent of the directed movement of substances between the cytoplasm and the nucleus.
negative regulation of protein import into nucleus
Any process that stops, prevents or reduces the frequency, rate or extent of the movement of proteins from the cytoplasm into the nucleus.
regulation of transcription factor import into nucleus
Any process that modulates the frequency, rate or extent of the movement of a transcription factor from the cytoplasm to the nucleus.
negative regulation of transcription factor import into nucleus
Any process that stops, prevents or reduces the frequency, rate or extent of the movement of a transcription factor from the cytoplasm to the nucleus.
protein import into nucleus
The directed movement of a protein from the cytoplasm to the nucleus.
negative regulation of transcription factor import into nucleus
Any process that stops, prevents or reduces the frequency, rate or extent of the movement of a transcription factor from the cytoplasm to the nucleus.


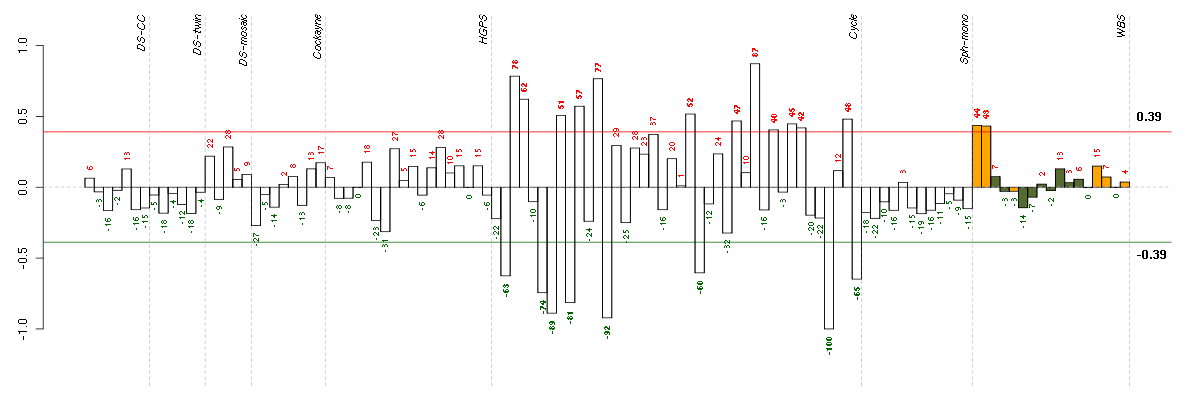
ABCD1ATP-binding cassette, sub-family D (ALD), member 1 (205142_x_at), score: 0.73 ABLIM1actin binding LIM protein 1 (200965_s_at), score: 0.78 ACSL4acyl-CoA synthetase long-chain family member 4 (202422_s_at), score: -0.84 ACTR3BARP3 actin-related protein 3 homolog B (yeast) (218868_at), score: -0.79 ADCK2aarF domain containing kinase 2 (221893_s_at), score: 0.83 AGPAT11-acylglycerol-3-phosphate O-acyltransferase 1 (lysophosphatidic acid acyltransferase, alpha) (215535_s_at), score: 0.81 ALDH1L1aldehyde dehydrogenase 1 family, member L1 (215798_at), score: -0.76 ARSAarylsulfatase A (204443_at), score: 0.72 ATF5activating transcription factor 5 (204999_s_at), score: 0.74 ATP2C1ATPase, Ca++ transporting, type 2C, member 1 (211137_s_at), score: -0.74 BAT2HLA-B associated transcript 2 (212081_x_at), score: 0.89 BCL2L1BCL2-like 1 (215037_s_at), score: 0.77 BTBD2BTB (POZ) domain containing 2 (207722_s_at), score: 0.89 C14orf94chromosome 14 open reading frame 94 (218383_at), score: 0.73 C4orf43chromosome 4 open reading frame 43 (218513_at), score: -0.85 C5orf44chromosome 5 open reading frame 44 (218674_at), score: -0.84 CALB2calbindin 2 (205428_s_at), score: -0.81 CALCOCO1calcium binding and coiled-coil domain 1 (209002_s_at), score: 0.9 CD248CD248 molecule, endosialin (219025_at), score: 0.73 CDC42BPBCDC42 binding protein kinase beta (DMPK-like) (217849_s_at), score: 0.87 CDC42EP4CDC42 effector protein (Rho GTPase binding) 4 (214721_x_at), score: 0.76 CHUKconserved helix-loop-helix ubiquitous kinase (209666_s_at), score: -0.79 CICcapicua homolog (Drosophila) (212784_at), score: 0.87 CIZ1CDKN1A interacting zinc finger protein 1 (205516_x_at), score: 0.81 CNOT3CCR4-NOT transcription complex, subunit 3 (203239_s_at), score: 0.91 CRTC3CREB regulated transcription coactivator 3 (218648_at), score: 0.86 CYP3A4cytochrome P450, family 3, subfamily A, polypeptide 4 (205998_x_at), score: -0.73 DDB2damage-specific DNA binding protein 2, 48kDa (203409_at), score: 0.72 DENND5BDENN/MADD domain containing 5B (215058_at), score: -0.73 DKFZP564O0823DKFZP564O0823 protein (204687_at), score: -0.73 DMPKdystrophia myotonica-protein kinase (37996_s_at), score: 0.77 DNAJB9DnaJ (Hsp40) homolog, subfamily B, member 9 (202843_at), score: -0.93 DNAJC6DnaJ (Hsp40) homolog, subfamily C, member 6 (204720_s_at), score: -0.74 DOPEY1dopey family member 1 (40612_at), score: -0.86 DSPPdentin sialophosphoprotein (221681_s_at), score: -0.73 ECM1extracellular matrix protein 1 (209365_s_at), score: 0.75 EDEM1ER degradation enhancer, mannosidase alpha-like 1 (203279_at), score: -0.72 EIF4Eeukaryotic translation initiation factor 4E (201436_at), score: -0.81 ENTPD1ectonucleoside triphosphate diphosphohydrolase 1 (209473_at), score: -0.77 EPHA4EPH receptor A4 (206114_at), score: -0.86 FAM18Bfamily with sequence similarity 18, member B (218446_s_at), score: -0.9 FBXL14F-box and leucine-rich repeat protein 14 (213145_at), score: 0.8 FOXC2forkhead box C2 (MFH-1, mesenchyme forkhead 1) (214520_at), score: 0.76 FOXK2forkhead box K2 (203064_s_at), score: 0.75 FUBP3far upstream element (FUSE) binding protein 3 (212824_at), score: -0.72 G3BP2GTPase activating protein (SH3 domain) binding protein 2 (208841_s_at), score: -0.73 GFOD1glucose-fructose oxidoreductase domain containing 1 (219821_s_at), score: -0.91 GGA3golgi associated, gamma adaptin ear containing, ARF binding protein 3 (209411_s_at), score: 0.73 GK3Pglycerol kinase 3 pseudogene (215966_x_at), score: -0.79 GOLT1Bgolgi transport 1 homolog B (S. cerevisiae) (218193_s_at), score: -0.75 H1FXH1 histone family, member X (204805_s_at), score: 0.83 HAB1B1 for mucin (215778_x_at), score: -0.78 HISPPD1histidine acid phosphatase domain containing 1 (203253_s_at), score: -0.76 HMGA1high mobility group AT-hook 1 (210457_x_at), score: 0.86 HNRNPH2heterogeneous nuclear ribonucleoprotein H2 (H') (201132_at), score: -0.75 HSD17B6hydroxysteroid (17-beta) dehydrogenase 6 homolog (mouse) (37512_at), score: -1 IDUAiduronidase, alpha-L- (205059_s_at), score: 0.78 IGHMBP2immunoglobulin mu binding protein 2 (215980_s_at), score: -0.74 ITGA1integrin, alpha 1 (214660_at), score: -0.73 JUNDjun D proto-oncogene (203751_x_at), score: 0.76 JUPjunction plakoglobin (201015_s_at), score: 0.88 KAL1Kallmann syndrome 1 sequence (205206_at), score: -0.99 KIAA0562KIAA0562 (204075_s_at), score: -0.74 KIAA1305KIAA1305 (220911_s_at), score: 0.81 KLHL23kelch-like 23 (Drosophila) (213610_s_at), score: -0.81 LARP4La ribonucleoprotein domain family, member 4 (214155_s_at), score: -0.86 LARSleucyl-tRNA synthetase (217810_x_at), score: -0.75 LLGL2lethal giant larvae homolog 2 (Drosophila) (203713_s_at), score: -0.77 LOC128192similar to peptidyl-Pro cis trans isomerase (217346_at), score: -0.72 LOC391132similar to hCG2041276 (216177_at), score: -0.81 LOC440434hypothetical protein FLJ11822 (215090_x_at), score: 0.81 LPGAT1lysophosphatidylglycerol acyltransferase 1 (202651_at), score: -0.78 LRP8low density lipoprotein receptor-related protein 8, apolipoprotein e receptor (205282_at), score: -0.91 MAP1Smicrotubule-associated protein 1S (218522_s_at), score: 0.79 MAP3K6mitogen-activated protein kinase kinase kinase 6 (219278_at), score: 0.79 MAPK7mitogen-activated protein kinase 7 (35617_at), score: 0.72 MAPKAPK3mitogen-activated protein kinase-activated protein kinase 3 (202788_at), score: 0.73 MDM2Mdm2 p53 binding protein homolog (mouse) (205386_s_at), score: 0.73 MINK1misshapen-like kinase 1 (zebrafish) (214246_x_at), score: 0.78 MMP10matrix metallopeptidase 10 (stromelysin 2) (205680_at), score: -0.79 MUC1mucin 1, cell surface associated (207847_s_at), score: 0.83 MYL4myosin, light chain 4, alkali; atrial, embryonic (210088_x_at), score: -0.8 NARG1LNMDA receptor regulated 1-like (219378_at), score: -0.8 NCKAP1NCK-associated protein 1 (217465_at), score: -0.76 NDPNorrie disease (pseudoglioma) (206022_at), score: 0.73 NEFMneurofilament, medium polypeptide (205113_at), score: -0.93 NF1neurofibromin 1 (211094_s_at), score: 0.8 NFATC4nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 4 (205897_at), score: 0.77 NFKBIAnuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha (201502_s_at), score: 0.84 NOL10nucleolar protein 10 (218591_s_at), score: -0.76 NOVA2neuro-oncological ventral antigen 2 (206477_s_at), score: -0.77 NUP160nucleoporin 160kDa (212709_at), score: -0.74 NUPL2nucleoporin like 2 (204003_s_at), score: -0.77 OAS32'-5'-oligoadenylate synthetase 3, 100kDa (218400_at), score: 0.74 OLFML2Bolfactomedin-like 2B (213125_at), score: 0.82 OLR1oxidized low density lipoprotein (lectin-like) receptor 1 (210004_at), score: -0.85 OPCMLopioid binding protein/cell adhesion molecule-like (214111_at), score: -0.72 OSTM1osteopetrosis associated transmembrane protein 1 (218196_at), score: -0.81 PAK1p21 protein (Cdc42/Rac)-activated kinase 1 (209615_s_at), score: 0.72 PARVBparvin, beta (204629_at), score: 0.78 PCDH9protocadherin 9 (219737_s_at), score: -0.96 PCDHGA11protocadherin gamma subfamily A, 11 (211876_x_at), score: 0.75 PCDHGA3protocadherin gamma subfamily A, 3 (216352_x_at), score: 0.76 PDE5Aphosphodiesterase 5A, cGMP-specific (206757_at), score: 0.76 PHF7PHD finger protein 7 (215622_x_at), score: -0.73 PIP5K1Cphosphatidylinositol-4-phosphate 5-kinase, type I, gamma (212518_at), score: 0.78 PKN1protein kinase N1 (202161_at), score: 0.79 PLAUplasminogen activator, urokinase (211668_s_at), score: 0.73 PLCD1phospholipase C, delta 1 (205125_at), score: 0.8 PNPLA2patatin-like phospholipase domain containing 2 (212705_x_at), score: 0.72 PRMT3protein arginine methyltransferase 3 (213320_at), score: -0.85 PXNpaxillin (211823_s_at), score: 0.87 RAB1ARAB1A, member RAS oncogene family (213440_at), score: -0.95 RBM15BRNA binding motif protein 15B (202689_at), score: 0.73 REREarginine-glutamic acid dipeptide (RE) repeats (200940_s_at), score: 0.82 RNF220ring finger protein 220 (219988_s_at), score: 0.88 RP5-1000E10.4suppressor of IKK epsilon (221705_s_at), score: -0.79 RPL18AP6ribosomal protein L18a pseudogene 6 (216383_at), score: -0.82 RPLP2P1ribosomal protein, large P2, pseudogene 1 (216490_x_at), score: -0.73 RPS17P5ribosomal protein S17 pseudogene 5 (216348_at), score: -0.92 RRP15ribosomal RNA processing 15 homolog (S. cerevisiae) (219037_at), score: -0.78 SAV1salvador homolog 1 (Drosophila) (218276_s_at), score: 0.76 SBNO2strawberry notch homolog 2 (Drosophila) (204166_at), score: 0.82 SCAMP4secretory carrier membrane protein 4 (213244_at), score: 0.92 SH3GLB2SH3-domain GRB2-like endophilin B2 (218813_s_at), score: 0.77 SIGIRRsingle immunoglobulin and toll-interleukin 1 receptor (TIR) domain (52940_at), score: 0.77 SLC25A13solute carrier family 25, member 13 (citrin) (203775_at), score: -0.88 SLC33A1solute carrier family 33 (acetyl-CoA transporter), member 1 (203165_s_at), score: -0.71 SLC4A7solute carrier family 4, sodium bicarbonate cotransporter, member 7 (209884_s_at), score: -0.87 SLC5A5solute carrier family 5 (sodium iodide symporter), member 5 (211123_at), score: -0.82 SLMO2slowmo homolog 2 (Drosophila) (217851_s_at), score: -0.75 SMG6Smg-6 homolog, nonsense mediated mRNA decay factor (C. elegans) (214940_s_at), score: 0.75 SOLHsmall optic lobes homolog (Drosophila) (204275_at), score: 0.79 ST20suppressor of tumorigenicity 20 (210242_x_at), score: -0.74 STIM1stromal interaction molecule 1 (202764_at), score: 0.72 TENC1tensin like C1 domain containing phosphatase (tensin 2) (212494_at), score: 0.8 TIAF1TGFB1-induced anti-apoptotic factor 1 (202039_at), score: 0.81 TICAM2toll-like receptor adaptor molecule 2 (214658_at), score: -0.95 TLE4transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila) (204872_at), score: -0.75 TRIM21tripartite motif-containing 21 (204804_at), score: 0.81 TRMT2ATRM2 tRNA methyltransferase 2 homolog A (S. cerevisiae) (91617_at), score: -0.79 TSC2tuberous sclerosis 2 (215735_s_at), score: 0.74 TSKUtsukushin (218245_at), score: 0.81 TUBA4Btubulin, alpha 4b (pseudogene) (207490_at), score: -0.86 USF2upstream transcription factor 2, c-fos interacting (202152_x_at), score: 0.72 VCPIP1valosin containing protein (p97)/p47 complex interacting protein 1 (219810_at), score: -0.73 VEGFBvascular endothelial growth factor B (203683_s_at), score: 0.96 VPS37Bvacuolar protein sorting 37 homolog B (S. cerevisiae) (221704_s_at), score: 0.81 WDR4WD repeat domain 4 (221632_s_at), score: -0.84 WDR6WD repeat domain 6 (217734_s_at), score: 0.77 ZCCHC10zinc finger, CCHC domain containing 10 (221193_s_at), score: -0.83
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-TABM-263-raw-cel-1515486371.cel | 37 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485891.cel | 13 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485771.cel | 7 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485811.cel | 9 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485751.cel | 6 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486431.cel | 40 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485671.cel | 2 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486091.cel | 23 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486251.cel | 31 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486311.cel | 34 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| 10590_WBS.CEL | 2 | 8 | WBS | hgu133plus2 | WBS | WBS 1 |
| 10358_WBS.CEL | 1 | 8 | WBS | hgu133plus2 | WBS | WBS 1 |
| E-TABM-263-raw-cel-1515486291.cel | 33 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486171.cel | 27 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486411.cel | 39 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485791.cel | 8 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486071.cel | 22 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485831.cel | 10 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485711.cel | 4 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485871.cel | 12 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485691.cel | 3 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486211.cel | 29 | 6 | Cycle | hgu133a2 | none | Cycle 1 |