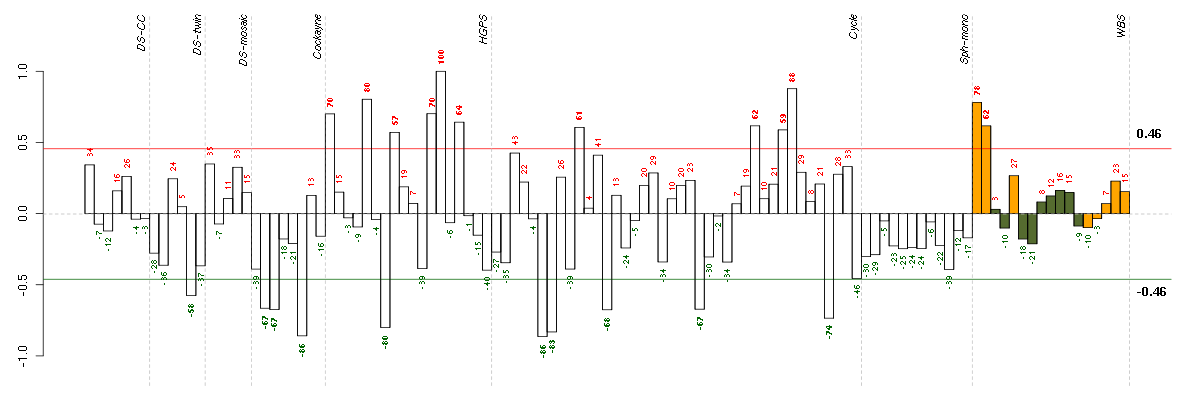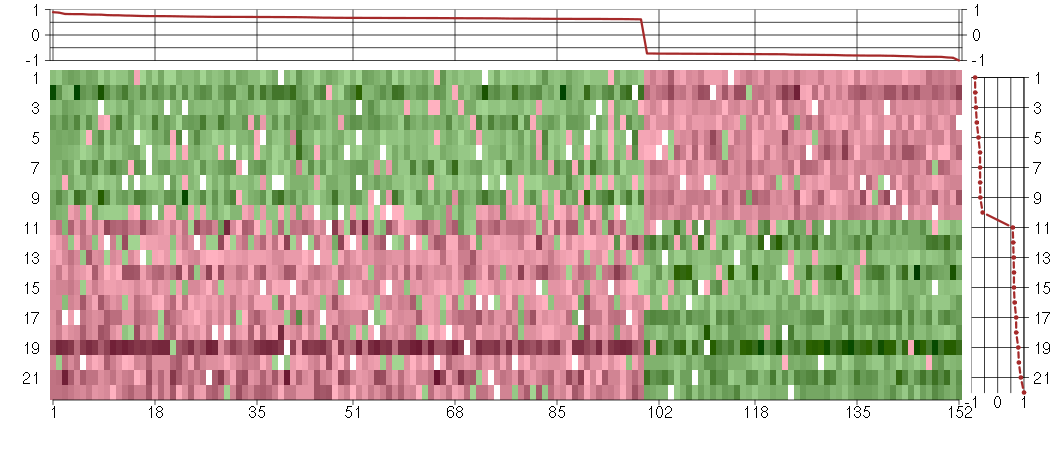

Under-expression is coded with green,
over-expression with red color.



molecular_function
Elemental activities, such as catalysis or binding, describing the actions of a gene product at the molecular level. A given gene product may exhibit one or more molecular functions.
catalytic activity
Catalysis of a biochemical reaction at physiological temperatures. In biologically catalyzed reactions, the reactants are known as substrates, and the catalysts are naturally occurring macromolecular substances known as enzymes. Enzymes possess specific binding sites for substrates, and are usually composed wholly or largely of protein, but RNA that has catalytic activity (ribozyme) is often also regarded as enzymatic.
lipase activity
Catalysis of the hydrolysis of a lipid or phospholipid.
hydrolase activity
Catalysis of the hydrolysis of various bonds, e.g. C-O, C-N, C-C, phosphoric anhydride bonds, etc. Hydrolase is the systematic name for any enzyme of EC class 3.
hydrolase activity, acting on ester bonds
Catalysis of the hydrolysis of any ester bond.
all
This term is the most general term possible
ABCD1ATP-binding cassette, sub-family D (ALD), member 1 (205142_x_at), score: 0.72 ABHD3abhydrolase domain containing 3 (213017_at), score: -0.86 ACN9ACN9 homolog (S. cerevisiae) (218981_at), score: -0.82 AESamino-terminal enhancer of split (217729_s_at), score: 0.66 AHNAK2AHNAK nucleoprotein 2 (212992_at), score: 0.71 AP2A2adaptor-related protein complex 2, alpha 2 subunit (212159_x_at), score: 0.69 APBB1amyloid beta (A4) precursor protein-binding, family B, member 1 (Fe65) (202652_at), score: 0.63 ARSAarylsulfatase A (204443_at), score: 0.88 ATP6AP1ATPase, H+ transporting, lysosomal accessory protein 1 (207809_s_at), score: 0.63 BCKDKbranched chain ketoacid dehydrogenase kinase (202030_at), score: 0.62 BNIP3LBCL2/adenovirus E1B 19kDa interacting protein 3-like (221479_s_at), score: 0.7 BTBD2BTB (POZ) domain containing 2 (207722_s_at), score: 0.82 C14orf106chromosome 14 open reading frame 106 (206500_s_at), score: -0.73 C20orf149chromosome 20 open reading frame 149 (218010_x_at), score: 0.64 C3orf18chromosome 3 open reading frame 18 (219114_at), score: 0.63 C4orf15chromosome 4 open reading frame 15 (210054_at), score: -0.73 C4orf31chromosome 4 open reading frame 31 (219747_at), score: 0.63 C4orf43chromosome 4 open reading frame 43 (218513_at), score: -0.8 CALCOCO1calcium binding and coiled-coil domain 1 (209002_s_at), score: 0.82 CAPGcapping protein (actin filament), gelsolin-like (201850_at), score: 0.71 CASP7caspase 7, apoptosis-related cysteine peptidase (207181_s_at), score: -0.74 CD248CD248 molecule, endosialin (219025_at), score: 0.82 CD81CD81 molecule (200675_at), score: 0.63 CDC7cell division cycle 7 homolog (S. cerevisiae) (204510_at), score: -0.73 CEBPZCCAAT/enhancer binding protein (C/EBP), zeta (203341_at), score: -0.75 CENPQcentromere protein Q (219294_at), score: -0.86 CHORDC1cysteine and histidine-rich domain (CHORD)-containing 1 (218566_s_at), score: -0.85 CHPFchondroitin polymerizing factor (202175_at), score: 0.75 CHUKconserved helix-loop-helix ubiquitous kinase (209666_s_at), score: -0.78 CICcapicua homolog (Drosophila) (212784_at), score: 0.7 CKBcreatine kinase, brain (200884_at), score: 0.75 CLIP3CAP-GLY domain containing linker protein 3 (212358_at), score: 0.66 COL8A2collagen, type VIII, alpha 2 (221900_at), score: 0.82 CRATcarnitine acetyltransferase (209522_s_at), score: 0.63 CREB3L1cAMP responsive element binding protein 3-like 1 (213059_at), score: 0.64 CRTC3CREB regulated transcription coactivator 3 (218648_at), score: 0.7 CSDC2cold shock domain containing C2, RNA binding (209981_at), score: 0.67 CUL7cullin 7 (36084_at), score: 0.72 DBF4DBF4 homolog (S. cerevisiae) (204244_s_at), score: -0.74 DBN1drebrin 1 (217025_s_at), score: 0.66 DDAH2dimethylarginine dimethylaminohydrolase 2 (215537_x_at), score: 0.64 DDR1discoidin domain receptor tyrosine kinase 1 (210749_x_at), score: 0.63 DHRS3dehydrogenase/reductase (SDR family) member 3 (202481_at), score: 0.62 ECM1extracellular matrix protein 1 (209365_s_at), score: 0.9 EFEMP2EGF-containing fibulin-like extracellular matrix protein 2 (209356_x_at), score: 0.67 EIF4Eeukaryotic translation initiation factor 4E (201436_at), score: -0.78 EPS8L2EPS8-like 2 (218180_s_at), score: 0.76 FAM29Afamily with sequence similarity 29, member A (218602_s_at), score: -0.81 FASNfatty acid synthase (212218_s_at), score: 0.63 FKBP8FK506 binding protein 8, 38kDa (208255_s_at), score: 0.77 GARTphosphoribosylglycinamide formyltransferase, phosphoribosylglycinamide synthetase, phosphoribosylaminoimidazole synthetase (210005_at), score: -0.8 GDPD5glycerophosphodiester phosphodiesterase domain containing 5 (32502_at), score: 0.67 GFOD1glucose-fructose oxidoreductase domain containing 1 (219821_s_at), score: -0.74 GINS3GINS complex subunit 3 (Psf3 homolog) (45633_at), score: -0.8 GLSglutaminase (203159_at), score: 0.61 GPR124G protein-coupled receptor 124 (221814_at), score: 0.62 GRINAglutamate receptor, ionotropic, N-methyl D-aspartate-associated protein 1 (glutamate binding) (212090_at), score: 0.77 GTF2F2general transcription factor IIF, polypeptide 2, 30kDa (209595_at), score: -0.78 GTF2H2general transcription factor IIH, polypeptide 2, 44kDa (221540_x_at), score: -0.74 GYS1glycogen synthase 1 (muscle) (201673_s_at), score: 0.66 HCFC1R1host cell factor C1 regulator 1 (XPO1 dependent) (45714_at), score: 0.67 HDAC5histone deacetylase 5 (202455_at), score: 0.8 HLA-Emajor histocompatibility complex, class I, E (200904_at), score: 0.63 HSPB6heat shock protein, alpha-crystallin-related, B6 (214767_s_at), score: 0.72 HSPE1heat shock 10kDa protein 1 (chaperonin 10) (205133_s_at), score: -0.81 INF2inverted formin, FH2 and WH2 domain containing (218144_s_at), score: 0.7 IRX5iroquois homeobox 5 (210239_at), score: 0.7 ISLRimmunoglobulin superfamily containing leucine-rich repeat (207191_s_at), score: 0.78 ISYNA1inositol-3-phosphate synthase 1 (222240_s_at), score: 0.66 JUNDjun D proto-oncogene (203751_x_at), score: 0.73 KLHL23kelch-like 23 (Drosophila) (213610_s_at), score: -0.76 LARP4La ribonucleoprotein domain family, member 4 (214155_s_at), score: -0.83 LBHlimb bud and heart development homolog (mouse) (221011_s_at), score: 0.73 LRRC40leucine rich repeat containing 40 (218577_at), score: -0.81 LSM5LSM5 homolog, U6 small nuclear RNA associated (S. cerevisiae) (202904_s_at), score: -0.78 LTBP3latent transforming growth factor beta binding protein 3 (219922_s_at), score: 0.71 MAPK3mitogen-activated protein kinase 3 (212046_x_at), score: 0.74 MEGF6multiple EGF-like-domains 6 (213942_at), score: 0.74 MFSD10major facilitator superfamily domain containing 10 (209215_at), score: 0.71 MGAT4Bmannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme B (220189_s_at), score: 0.67 MICBMHC class I polypeptide-related sequence B (206247_at), score: -0.75 MUC1mucin 1, cell surface associated (207847_s_at), score: 0.63 MXRA8matrix-remodelling associated 8 (213422_s_at), score: 0.65 NBL1neuroblastoma, suppression of tumorigenicity 1 (37005_at), score: 0.68 NDUFA5NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 5, 13kDa (201304_at), score: -0.73 NUCB1nucleobindin 1 (200646_s_at), score: 0.66 NUP160nucleoporin 160kDa (212709_at), score: -0.85 OBSL1obscurin-like 1 (212775_at), score: 0.64 ORAI3ORAI calcium release-activated calcium modulator 3 (221864_at), score: 0.7 ORC6Lorigin recognition complex, subunit 6 like (yeast) (219105_x_at), score: -0.73 PCBP4poly(rC) binding protein 4 (209361_s_at), score: 0.65 PCDHG@protocadherin gamma cluster (215836_s_at), score: 0.67 PCDHGA1protocadherin gamma subfamily A, 1 (209079_x_at), score: 0.71 PCDHGC3protocadherin gamma subfamily C, 3 (211066_x_at), score: 0.67 PDK2pyruvate dehydrogenase kinase, isozyme 2 (202590_s_at), score: 0.66 PIP5K1Cphosphatidylinositol-4-phosphate 5-kinase, type I, gamma (212518_at), score: 0.67 PLA2G4Aphospholipase A2, group IVA (cytosolic, calcium-dependent) (210145_at), score: -0.74 PLCD1phospholipase C, delta 1 (205125_at), score: 0.76 PLD3phospholipase D family, member 3 (201050_at), score: 0.74 PLEC1plectin 1, intermediate filament binding protein 500kDa (216971_s_at), score: 0.74 PLSCR3phospholipid scramblase 3 (218828_at), score: 0.67 PNPLA2patatin-like phospholipase domain containing 2 (212705_x_at), score: 0.64 PNPLA6patatin-like phospholipase domain containing 6 (203718_at), score: 0.63 PNRC1proline-rich nuclear receptor coactivator 1 (209034_at), score: 0.66 POLR3Gpolymerase (RNA) III (DNA directed) polypeptide G (32kD) (206653_at), score: -0.77 PPAP2Aphosphatidic acid phosphatase type 2A (209147_s_at), score: 0.65 PPATphosphoribosyl pyrophosphate amidotransferase (209433_s_at), score: -0.86 PRMT3protein arginine methyltransferase 3 (213320_at), score: -0.74 PRPF38BPRP38 pre-mRNA processing factor 38 (yeast) domain containing B (218040_at), score: -0.81 PRPS2phosphoribosyl pyrophosphate synthetase 2 (203401_at), score: -0.88 RAB11BRAB11B, member RAS oncogene family (34478_at), score: 0.66 RBL1retinoblastoma-like 1 (p107) (205296_at), score: -0.75 RFNGRFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase (212968_at), score: 0.67 RNF187ring finger protein 187 (212155_at), score: 0.71 RPLP2P1ribosomal protein, large P2, pseudogene 1 (216490_x_at), score: -0.74 RPS17P5ribosomal protein S17 pseudogene 5 (216348_at), score: -0.75 RRP15ribosomal RNA processing 15 homolog (S. cerevisiae) (219037_at), score: -0.81 SDC3syndecan 3 (202898_at), score: 0.68 SERPING1serpin peptidase inhibitor, clade G (C1 inhibitor), member 1 (200986_at), score: 0.74 SIP1survival of motor neuron protein interacting protein 1 (205063_at), score: -0.83 SLC22A18solute carrier family 22, member 18 (204981_at), score: 0.64 SLC25A1solute carrier family 25 (mitochondrial carrier; citrate transporter), member 1 (210010_s_at), score: 0.64 SLC25A13solute carrier family 25, member 13 (citrin) (203775_at), score: -1 SLC25A17solute carrier family 25 (mitochondrial carrier; peroxisomal membrane protein, 34kDa), member 17 (211754_s_at), score: -0.75 SLC6A8solute carrier family 6 (neurotransmitter transporter, creatine), member 8 (202219_at), score: 0.67 SMCHD1structural maintenance of chromosomes flexible hinge domain containing 1 (212569_at), score: -0.89 SMPD1sphingomyelin phosphodiesterase 1, acid lysosomal (209420_s_at), score: 0.67 SNTA1syntrophin, alpha 1 (dystrophin-associated protein A1, 59kDa, acidic component) (203516_at), score: 0.67 SORBS3sorbin and SH3 domain containing 3 (209253_at), score: 0.65 SP3Sp3 transcription factor (213168_at), score: -0.79 SPATA20spermatogenesis associated 20 (218164_at), score: 0.71 SREBF2sterol regulatory element binding transcription factor 2 (201247_at), score: 0.71 SUSD5sushi domain containing 5 (214954_at), score: -0.77 SUZ12suppressor of zeste 12 homolog (Drosophila) (212287_at), score: -0.78 TBC1D17TBC1 domain family, member 17 (218466_at), score: 0.69 TCERG1transcription elongation regulator 1 (202396_at), score: -0.82 TENC1tensin like C1 domain containing phosphatase (tensin 2) (212494_at), score: 0.67 TFAMtranscription factor A, mitochondrial (203177_x_at), score: -0.75 TIMP2TIMP metallopeptidase inhibitor 2 (203167_at), score: 0.8 TMEM8transmembrane protein 8 (five membrane-spanning domains) (221882_s_at), score: 0.71 TOPBP1topoisomerase (DNA) II binding protein 1 (202633_at), score: -0.73 TSC2tuberous sclerosis 2 (215735_s_at), score: 0.71 TTF2transcription termination factor, RNA polymerase II (204407_at), score: -0.73 UBA2ubiquitin-like modifier activating enzyme 2 (201177_s_at), score: -0.77 UBE2V2ubiquitin-conjugating enzyme E2 variant 2 (209096_at), score: -0.73 UBXN6UBX domain protein 6 (220757_s_at), score: 0.71 USF2upstream transcription factor 2, c-fos interacting (202152_x_at), score: 0.67 VAT1vesicle amine transport protein 1 homolog (T. californica) (208626_s_at), score: 0.68 VEGFBvascular endothelial growth factor B (203683_s_at), score: 0.8 WBP2WW domain binding protein 2 (209117_at), score: 0.63 WDHD1WD repeat and HMG-box DNA binding protein 1 (216228_s_at), score: -0.74 ZNF107zinc finger protein 107 (205739_x_at), score: -0.75
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-TABM-263-raw-cel-1515485751.cel | 6 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-GEOD-3407-raw-cel-1437949750.cel | 6 | 4 | Cockayne | hgu133a | CS | CSB |
| E-TABM-263-raw-cel-1515485771.cel | 7 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-GEOD-3860-raw-cel-1561690272.cel | 7 | 5 | HGPS | hgu133a | HGPS | AG11498 |
| E-TABM-263-raw-cel-1515486371.cel | 37 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485891.cel | 13 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-GEOD-3407-raw-cel-1437949655.cel | 3 | 4 | Cockayne | hgu133a | none | CSB |
| E-TABM-263-raw-cel-1515486091.cel | 23 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-GEOD-3407-raw-cel-1437949579.cel | 2 | 4 | Cockayne | hgu133a | none | CSB |
| 5CTwin.CEL | 5 | 2 | DS-twin | hgu133plus2 | Down | DS-twin 5 |
| E-GEOD-3860-raw-cel-1561690304.cel | 8 | 5 | HGPS | hgu133a | none | GMO8398C |
| E-TABM-263-raw-cel-1515486271.cel | 32 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485831.cel | 10 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| 10590_WBS.CEL | 2 | 8 | WBS | hgu133plus2 | WBS | WBS 1 |
| E-TABM-263-raw-cel-1515486211.cel | 29 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-GEOD-3860-raw-cel-1561690416.cel | 15 | 5 | HGPS | hgu133a | none | GM0316B |
| E-GEOD-3860-raw-cel-1561690199.cel | 1 | 5 | HGPS | hgu133a | none | GM0316B |
| E-GEOD-3860-raw-cel-1561690360.cel | 12 | 5 | HGPS | hgu133a | none | GM0316B |
| 10358_WBS.CEL | 1 | 8 | WBS | hgu133plus2 | WBS | WBS 1 |
| E-GEOD-3860-raw-cel-1561690248.cel | 5 | 5 | HGPS | hgu133a | HGPS | AG11513 |
| E-TABM-263-raw-cel-1515486291.cel | 33 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-GEOD-3860-raw-cel-1561690376.cel | 13 | 5 | HGPS | hgu133a | HGPS | AG11513 |