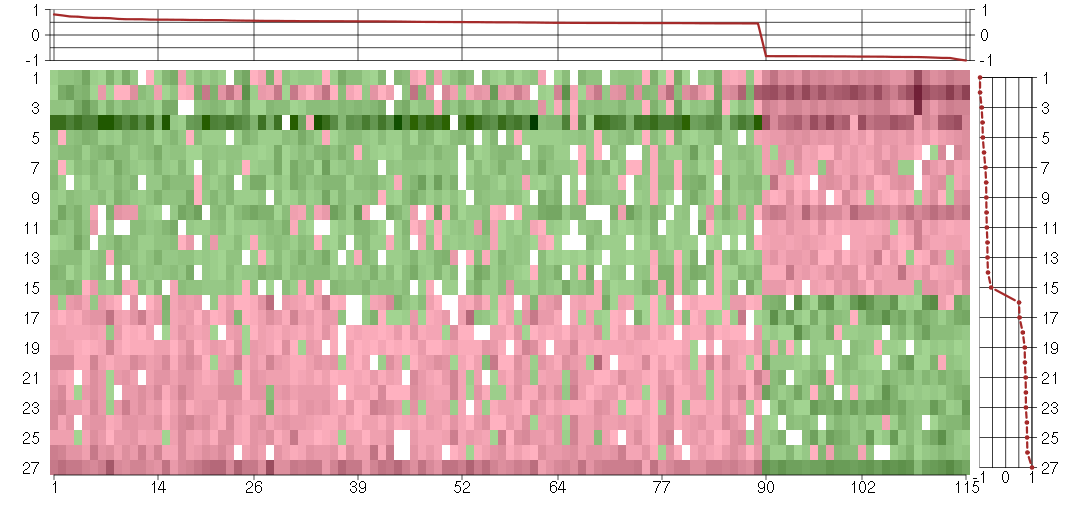

Under-expression is coded with green,
over-expression with red color.

immune system process
Any process involved in the development or functioning of the immune system, an organismal system for calibrated responses to potential internal or invasive threats.
antigen processing and presentation of peptide antigen via MHC class I
The process by which an antigen-presenting cell expresses a peptide antigen on its cell surface in association with an MHC class I protein complex. Class I here refers to classical class I molecules.
multicellular organismal development
The biological process whose specific outcome is the progression of a multicellular organism over time from an initial condition (e.g. a zygote or a young adult) to a later condition (e.g. a multicellular animal or an aged adult).
biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
antigen processing and presentation
The process by which an antigen-presenting cell expresses antigen (peptide or lipid) on its cell surface in association with an MHC protein complex.
multicellular organismal process
Any biological process, occurring at the level of a multicellular organism, pertinent to its function.
developmental process
A biological process whose specific outcome is the progression of an integrated living unit: an anatomical structure (which may be a subcellular structure, cell, tissue, or organ), or organism over time from an initial condition to a later condition.
antigen processing and presentation of peptide antigen
The process by which an antigen-presenting cell expresses peptide antigen in association with an MHC protein complex on its cell surface, including proteolysis and transport steps for the peptide antigen both prior to and following assembly with the MHC protein complex. The peptide antigen is typically, but not always, processed from an endogenous or exogenous protein.
system development
The process whose specific outcome is the progression of an organismal system over time, from its formation to the mature structure. A system is a regularly interacting or interdependent group of organs or tissues that work together to carry out a given biological process.
anatomical structure development
The biological process whose specific outcome is the progression of an anatomical structure from an initial condition to its mature state. This process begins with the formation of the structure and ends with the mature structure, whatever form that may be including its natural destruction. An anatomical structure is any biological entity that occupies space and is distinguished from its surroundings. Anatomical structures can be macroscopic such as a carpel, or microscopic such as an acrosome.
regulation of biological process
Any process that modulates the frequency, rate or extent of a biological process. Biological processes are regulated by many means; examples include the control of gene expression, protein modification or interaction with a protein or substrate molecule.
regulation of multicellular organismal process
Any process that modulates the frequency, rate or extent of a multicellular organismal process, the processes pertinent to the function of a multicellular organism above the cellular level; includes the integrated processes of tissues and organs.
biological regulation
Any process that modulates the frequency, rate or extent of any biological process, quality or function.
all
This term is the most general term possible
multicellular organismal development
The biological process whose specific outcome is the progression of a multicellular organism over time from an initial condition (e.g. a zygote or a young adult) to a later condition (e.g. a multicellular animal or an aged adult).
regulation of multicellular organismal process
Any process that modulates the frequency, rate or extent of a multicellular organismal process, the processes pertinent to the function of a multicellular organism above the cellular level; includes the integrated processes of tissues and organs.
regulation of biological process
Any process that modulates the frequency, rate or extent of a biological process. Biological processes are regulated by many means; examples include the control of gene expression, protein modification or interaction with a protein or substrate molecule.
system development
The process whose specific outcome is the progression of an organismal system over time, from its formation to the mature structure. A system is a regularly interacting or interdependent group of organs or tissues that work together to carry out a given biological process.

plasma membrane
The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins.
membrane
Double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
extracellular region
The space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers the host cell environment outside an intracellular parasite.
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
cell
The basic structural and functional unit of all organisms. Includes the plasma membrane and any external encapsulating structures such as the cell wall and cell envelope.
external side of plasma membrane
The side of the plasma membrane that is opposite to the side that faces the cytoplasm.
cell surface
The external part of the cell wall and/or plasma membrane.
extracellular region part
Any constituent part of the extracellular region, the space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers constituent parts of the host cell environment outside an intracellular parasite.
membrane part
Any constituent part of a membrane, a double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
plasma membrane part
Any constituent part of the plasma membrane, the membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins.
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
all
This term is the most general term possible
extracellular region part
Any constituent part of the extracellular region, the space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers constituent parts of the host cell environment outside an intracellular parasite.
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
membrane part
Any constituent part of a membrane, a double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
plasma membrane part
Any constituent part of the plasma membrane, the membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins.
external side of plasma membrane
The side of the plasma membrane that is opposite to the side that faces the cytoplasm.

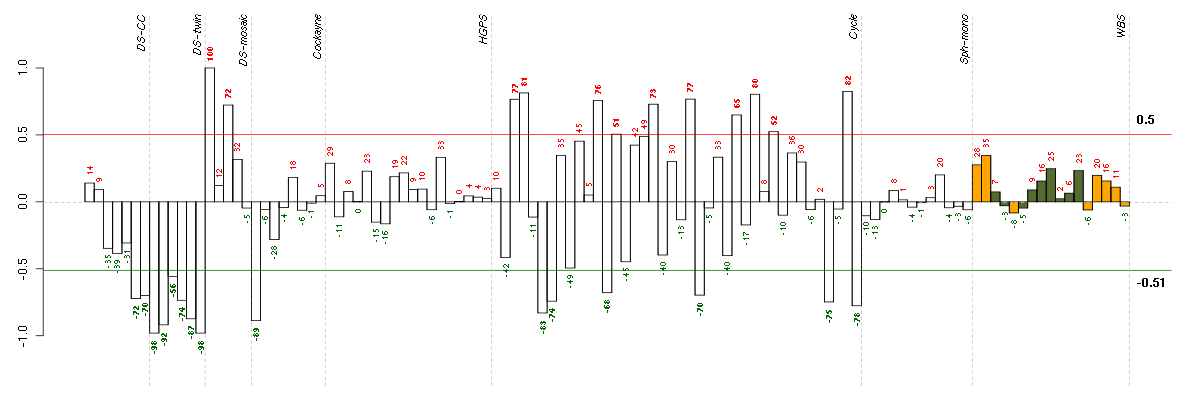
ABLIM3actin binding LIM protein family, member 3 (205730_s_at), score: 0.5 ACTN3actinin, alpha 3 (206891_at), score: -0.83 AGRNagrin (217419_x_at), score: 0.54 AKIRIN1akirin 1 (217893_s_at), score: 0.54 ARSAarylsulfatase A (204443_at), score: 0.62 BAT2HLA-B associated transcript 2 (212081_x_at), score: 0.57 BAT2LHLA-B associated transcript 2-like (212068_s_at), score: 0.55 C16orf45chromosome 16 open reading frame 45 (212736_at), score: 0.46 C9orf167chromosome 9 open reading frame 167 (219620_x_at), score: 0.53 CDAcytidine deaminase (205627_at), score: 0.47 CDADC1cytidine and dCMP deaminase domain containing 1 (221015_s_at), score: -0.83 CDC42SE1CDC42 small effector 1 (218157_x_at), score: 0.46 COL11A2collagen, type XI, alpha 2 (216993_s_at), score: -0.83 COL18A1collagen, type XVIII, alpha 1 (209081_s_at), score: 0.54 CST3cystatin C (201360_at), score: 0.51 DESdesmin (202222_s_at), score: -0.86 DGCR2DiGeorge syndrome critical region gene 2 (214198_s_at), score: -0.86 DOK4docking protein 4 (209691_s_at), score: 0.5 DOPEY1dopey family member 1 (40612_at), score: -0.86 DRG2developmentally regulated GTP binding protein 2 (203267_s_at), score: 0.46 DSPPdentin sialophosphoprotein (221681_s_at), score: -0.85 DUS2Ldihydrouridine synthase 2-like, SMM1 homolog (S. cerevisiae) (47105_at), score: -0.83 DUSP3dual specificity phosphatase 3 (201537_s_at), score: 0.54 DUSP5dual specificity phosphatase 5 (209457_at), score: 0.47 ECE1endothelin converting enzyme 1 (201750_s_at), score: 0.47 ECM1extracellular matrix protein 1 (209365_s_at), score: 0.6 FBXO2F-box protein 2 (219305_x_at), score: -0.89 FCER1GFc fragment of IgE, high affinity I, receptor for; gamma polypeptide (204232_at), score: -0.84 FHOD1formin homology 2 domain containing 1 (218530_at), score: 0.77 FLJ40113golgi autoantigen, golgin subfamily a-like pseudogene (213212_x_at), score: -0.85 FOXC2forkhead box C2 (MFH-1, mesenchyme forkhead 1) (214520_at), score: 0.59 FOXF1forkhead box F1 (205935_at), score: 0.47 FOXK2forkhead box K2 (203064_s_at), score: 0.54 FURINfurin (paired basic amino acid cleaving enzyme) (201945_at), score: 0.62 GNL3LPguanine nucleotide binding protein-like 3 (nucleolar)-like pseudogene (220716_at), score: -0.87 GPX3glutathione peroxidase 3 (plasma) (214091_s_at), score: 0.55 HAB1B1 for mucin (215778_x_at), score: -0.86 HAMPhepcidin antimicrobial peptide (220491_at), score: -0.84 HBEGFheparin-binding EGF-like growth factor (203821_at), score: 0.48 HBS1LHBS1-like (S. cerevisiae) (209314_s_at), score: -0.83 HHEXhematopoietically expressed homeobox (215933_s_at), score: 0.53 HIST1H4Dhistone cluster 1, H4d (208076_at), score: -0.85 HLA-Cmajor histocompatibility complex, class I, C (211799_x_at), score: 0.46 HLA-Emajor histocompatibility complex, class I, E (200904_at), score: 0.69 HLA-Fmajor histocompatibility complex, class I, F (204806_x_at), score: 0.47 HPCAL1hippocalcin-like 1 (205462_s_at), score: 0.55 HSF1heat shock transcription factor 1 (202344_at), score: 0.58 IDUAiduronidase, alpha-L- (205059_s_at), score: 0.62 IGHMBP2immunoglobulin mu binding protein 2 (215980_s_at), score: -0.83 IL6STinterleukin 6 signal transducer (gp130, oncostatin M receptor) (204864_s_at), score: 0.47 INPP1inositol polyphosphate-1-phosphatase (202794_at), score: 0.54 JUNDjun D proto-oncogene (203751_x_at), score: 0.51 JUPjunction plakoglobin (201015_s_at), score: 0.45 KLHL26kelch-like 26 (Drosophila) (219354_at), score: 0.57 LEPREL2leprecan-like 2 (204854_at), score: 0.5 LIFleukemia inhibitory factor (cholinergic differentiation factor) (205266_at), score: 0.5 LOC100128809similar to hCG2045829 (215707_s_at), score: 0.47 LOC440434hypothetical protein FLJ11822 (215090_x_at), score: 0.48 MAGOH2mago-nashi homolog 2, proliferation-associated (Drosophila) (217693_x_at), score: -0.84 MAN1C1mannosidase, alpha, class 1C, member 1 (218918_at), score: 0.54 MAP1Smicrotubule-associated protein 1S (218522_s_at), score: 0.81 MAP3K6mitogen-activated protein kinase kinase kinase 6 (219278_at), score: 0.47 NACAP1nascent-polypeptide-associated complex alpha polypeptide pseudogene 1 (211445_x_at), score: -0.9 NFKB1nuclear factor of kappa light polypeptide gene enhancer in B-cells 1 (209239_at), score: 0.72 NID1nidogen 1 (202008_s_at), score: 0.59 NINJ1ninjurin 1 (203045_at), score: 0.6 NOTCH2Notch homolog 2 (Drosophila) (210756_s_at), score: 0.48 PCDHG@protocadherin gamma cluster (215836_s_at), score: 0.6 PCDHGA11protocadherin gamma subfamily A, 11 (211876_x_at), score: 0.56 PCDHGA3protocadherin gamma subfamily A, 3 (216352_x_at), score: 0.46 PDE5Aphosphodiesterase 5A, cGMP-specific (206757_at), score: 0.51 PHF7PHD finger protein 7 (215622_x_at), score: -0.9 PIP5K1Cphosphatidylinositol-4-phosphate 5-kinase, type I, gamma (212518_at), score: 0.67 PKP2plakophilin 2 (207717_s_at), score: 0.66 PLAUplasminogen activator, urokinase (211668_s_at), score: 0.55 PLK3polo-like kinase 3 (Drosophila) (204958_at), score: 0.46 POLSpolymerase (DNA directed) sigma (202466_at), score: 0.47 POM121POM121 membrane glycoprotein (rat) (212178_s_at), score: 0.48 PRKCDprotein kinase C, delta (202545_at), score: 0.53 PTPROprotein tyrosine phosphatase, receptor type, O (211600_at), score: 0.51 PXNpaxillin (211823_s_at), score: 0.6 RIPK1receptor (TNFRSF)-interacting serine-threonine kinase 1 (209941_at), score: 0.53 RNF130ring finger protein 130 (217865_at), score: 0.51 RNF24ring finger protein 24 (204669_s_at), score: 0.47 RPS17P5ribosomal protein S17 pseudogene 5 (216348_at), score: -1 RPS4Y1ribosomal protein S4, Y-linked 1 (201909_at), score: 0.47 SAV1salvador homolog 1 (Drosophila) (218276_s_at), score: 0.47 SCAMP4secretory carrier membrane protein 4 (213244_at), score: 0.52 SCAMP5secretory carrier membrane protein 5 (212699_at), score: -0.84 SCG2secretogranin II (chromogranin C) (204035_at), score: 0.49 SHBSrc homology 2 domain containing adaptor protein B (204657_s_at), score: 0.53 SIGLEC1sialic acid binding Ig-like lectin 1, sialoadhesin (44673_at), score: -0.85 SIN3BSIN3 homolog B, transcription regulator (yeast) (209352_s_at), score: 0.51 SLC2A8solute carrier family 2 (facilitated glucose transporter), member 8 (218985_at), score: 0.47 SLC9A3R2solute carrier family 9 (sodium/hydrogen exchanger), member 3 regulator 2 (209830_s_at), score: 0.58 SMG6Smg-6 homolog, nonsense mediated mRNA decay factor (C. elegans) (214940_s_at), score: 0.46 SOLHsmall optic lobes homolog (Drosophila) (204275_at), score: 0.6 SPATA20spermatogenesis associated 20 (218164_at), score: 0.5 SPRY4sprouty homolog 4 (Drosophila) (221489_s_at), score: 0.73 STOMstomatin (201060_x_at), score: 0.54 SYT1synaptotagmin I (203999_at), score: 0.49 TAPBPTAP binding protein (tapasin) (208829_at), score: 0.55 TERF2telomeric repeat binding factor 2 (203611_at), score: 0.5 TGFBR2transforming growth factor, beta receptor II (70/80kDa) (207334_s_at), score: 0.48 TLE6transducin-like enhancer of split 6 (E(sp1) homolog, Drosophila) (222219_s_at), score: -0.85 TMCC2transmembrane and coiled-coil domain family 2 (213096_at), score: -0.83 TNFAIP2tumor necrosis factor, alpha-induced protein 2 (202510_s_at), score: 0.52 TP53I11tumor protein p53 inducible protein 11 (214667_s_at), score: 0.58 TPD52tumor protein D52 (201690_s_at), score: 0.5 TPP1tripeptidyl peptidase I (200742_s_at), score: 0.56 TUBA4Btubulin, alpha 4b (pseudogene) (207490_at), score: -0.96 VEGFBvascular endothelial growth factor B (203683_s_at), score: 0.67 VPS37Bvacuolar protein sorting 37 homolog B (S. cerevisiae) (221704_s_at), score: 0.63 WDR6WD repeat domain 6 (217734_s_at), score: 0.46 ZNF282zinc finger protein 282 (212892_at), score: 0.47
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| 6Twin.CEL | 6 | 2 | DS-twin | hgu133plus2 | none | DS-twin 6 |
| 1Twin.CEL | 1 | 2 | DS-twin | hgu133plus2 | Down | DS-twin 1 |
| 2Twin.CEL | 2 | 2 | DS-twin | hgu133plus2 | none | DS-twin 2 |
| E-GEOD-3407-raw-cel-1437949557.cel | 1 | 4 | Cockayne | hgu133a | CS | eGFP |
| 5CTwin.CEL | 5 | 2 | DS-twin | hgu133plus2 | Down | DS-twin 5 |
| E-TABM-263-raw-cel-1515485751.cel | 6 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486431.cel | 40 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486371.cel | 37 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485771.cel | 7 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| 4Twin.CEL | 4 | 2 | DS-twin | hgu133plus2 | none | DS-twin 4 |
| t21c 08-03.CEL | 6 | 1 | DS-CC | hgu133a | Down | DS-CC 6 |
| t21d 08-03.CEL | 7 | 1 | DS-CC | hgu133a | Down | DS-CC 7 |
| E-TABM-263-raw-cel-1515486091.cel | 23 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485891.cel | 13 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| 3Twin.CEL | 3 | 2 | DS-twin | hgu133plus2 | Down | DS-twin 3 |
| E-TABM-263-raw-cel-1515485911.cel | 14 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486251.cel | 31 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486171.cel | 27 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| 46C.CEL | 3 | 3 | DS-mosaic | hgu133plus2 | none | DS-mosaic 3 |
| E-TABM-263-raw-cel-1515485991.cel | 18 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485871.cel | 12 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485691.cel | 3 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486071.cel | 22 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486211.cel | 29 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485711.cel | 4 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486411.cel | 39 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| 46A.CEL | 1 | 3 | DS-mosaic | hgu133plus2 | none | DS-mosaic 1 |