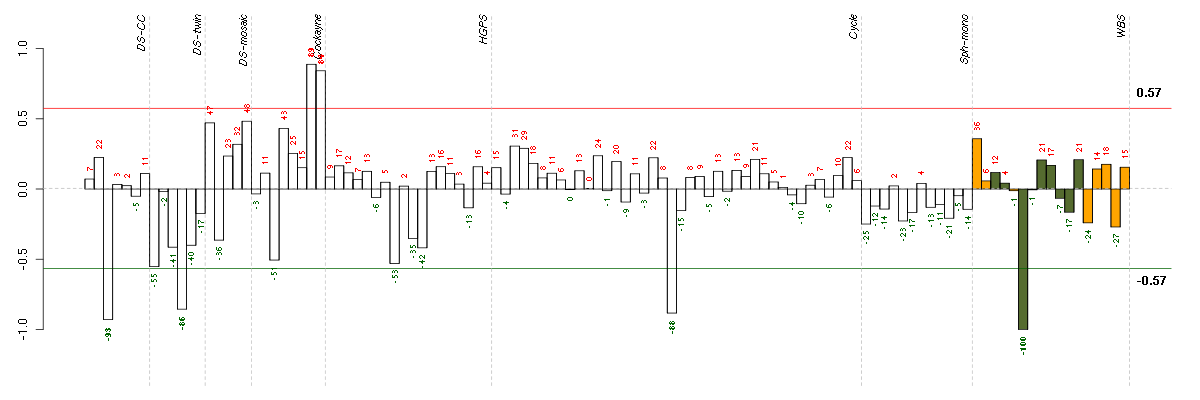

Under-expression is coded with green,
over-expression with red color.

MAPKKK cascade
Cascade of at least three protein kinase activities culminating in the phosphorylation and activation of a MAP kinase. MAPKKK cascades lie downstream of numerous signaling pathways.
activation of MAPK activity
The initiation of the activity of the inactive enzyme MAP kinase by phosphorylation by a MAPKK.
metabolic process
The chemical reactions and pathways, including anabolism and catabolism, by which living organisms transform chemical substances. Metabolic processes typically transform small molecules, but also include macromolecular processes such as DNA repair and replication, and protein synthesis and degradation.
signal transduction
The cascade of processes by which a signal interacts with a receptor, causing a change in the level or activity of a second messenger or other downstream target, and ultimately effecting a change in the functioning of the cell.
protein modification process
The covalent alteration of one or more amino acids occurring in proteins, peptides and nascent polypeptides (co-translational, post-translational modifications). Includes the modification of charged tRNAs that are destined to occur in a protein (pre-translation modification).
protein amino acid phosphorylation
The process of introducing a phosphate group on to a protein.
phosphorus metabolic process
The chemical reactions and pathways involving the nonmetallic element phosphorus or compounds that contain phosphorus, usually in the form of a phosphate group (PO4).
phosphate metabolic process
The chemical reactions and pathways involving the phosphate group, the anion or salt of any phosphoric acid.
cell communication
Any process that mediates interactions between a cell and its surroundings. Encompasses interactions such as signaling or attachment between one cell and another cell, between a cell and an extracellular matrix, or between a cell and any other aspect of its environment.
intracellular signaling cascade
A series of reactions within the cell that occur as a result of a single trigger reaction or compound.
protein kinase cascade
A series of reactions, mediated by protein kinases, which occurs as a result of a single trigger reaction or compound.
small GTPase mediated signal transduction
Any series of molecular signals in which a small monomeric GTPase relays one or more of the signals.
Ras protein signal transduction
A series of molecular signals within the cell that are mediated by a member of the Ras superfamily of proteins switching to a GTP-bound active state.
biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
cellular process
Any process that is carried out at the cellular level, but not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
phosphorylation
The process of introducing a phosphate group into a molecule, usually with the formation of a phosphoric ester, a phosphoric anhydride or a phosphoric amide.
protein metabolic process
The chemical reactions and pathways involving a specific protein, rather than of proteins in general. Includes protein modification.
positive regulation of kinase activity
Any process that activates or increases the frequency, rate or extent of kinase activity, the catalysis of the transfer of a phosphate group, usually from ATP, to a substrate molecule.
cellular biopolymer metabolic process
The chemical reactions and pathways involving biopolymers, long, repeating chains of monomers found in nature, such as polysaccharides and proteins, as carried out by individual cells.
positive regulation of catalytic activity
Any process that activates or increases the activity of an enzyme.
macromolecule metabolic process
The chemical reactions and pathways involving macromolecules, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass.
biopolymer metabolic process
The chemical reactions and pathways involving biopolymers, long, repeating chains of monomers found in nature, such as polysaccharides and proteins.
regulation of MAP kinase activity
Any process that modulates the frequency, rate or extent of MAP kinase activity.
positive regulation of MAP kinase activity
Any process that activates or increases the frequency, rate or extent of MAP kinase activity.
biopolymer modification
The covalent alteration of one or more monomeric units in a polypeptide, polynucleotide, polysaccharide, or other biological polymer, resulting in a change in its properties.
regulation of kinase activity
Any process that modulates the frequency, rate or extent of kinase activity, the catalysis of the transfer of a phosphate group, usually from ATP, to a substrate molecule.
post-translational protein modification
The covalent alteration of one or more amino acids occurring in a protein after the protein has been completely translated and released from the ribosome.
cellular metabolic process
The chemical reactions and pathways by which individual cells transform chemical substances.
primary metabolic process
The chemical reactions and pathways involving those compounds which are formed as a part of the normal anabolic and catabolic processes. These processes take place in most, if not all, cells of the organism.
cellular macromolecule metabolic process
The chemical reactions and pathways involving macromolecules, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass, as carried out by individual cells.
cellular protein metabolic process
The chemical reactions and pathways involving a specific protein, rather than of proteins in general, occurring at the level of an individual cell. Includes protein modification.
regulation of protein kinase activity
Any process that modulates the frequency, rate or extent of protein kinase activity.
positive regulation of protein kinase activity
Any process that activates or increases the frequency, rate or extent of protein kinase activity.
regulation of biological process
Any process that modulates the frequency, rate or extent of a biological process. Biological processes are regulated by many means; examples include the control of gene expression, protein modification or interaction with a protein or substrate molecule.
regulation of catalytic activity
Any process that modulates the activity of an enzyme.
regulation of cellular process
Any process that modulates the frequency, rate or extent of a cellular process, any of those that are carried out at the cellular level, but are not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
regulation of transferase activity
Any process that modulates the frequency, rate or extent of transferase activity, the catalysis of the transfer of a group, e.g. a methyl group, glycosyl group, acyl group, phosphorus-containing, or other groups, from one compound (generally regarded as the donor) to another compound (generally regarded as the acceptor). Transferase is the systematic name for any enzyme of EC class 2.
positive regulation of transferase activity
Any process that activates or increases the frequency, rate or extent of transferase activity, the catalysis of the transfer of a group, e.g. a methyl group, glycosyl group, acyl group, phosphorus-containing, or other groups, from a donor compound to an acceptor.
biological regulation
Any process that modulates the frequency, rate or extent of any biological process, quality or function.
regulation of molecular function
Any process that modulates the frequency, rate or extent of molecular functions. Molecular functions are elemental biological activities occurring at the molecular level, such as catalysis or binding.
all
This term is the most general term possible
cellular metabolic process
The chemical reactions and pathways by which individual cells transform chemical substances.
regulation of cellular process
Any process that modulates the frequency, rate or extent of a cellular process, any of those that are carried out at the cellular level, but are not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
regulation of biological process
Any process that modulates the frequency, rate or extent of a biological process. Biological processes are regulated by many means; examples include the control of gene expression, protein modification or interaction with a protein or substrate molecule.
cellular macromolecule metabolic process
The chemical reactions and pathways involving macromolecules, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass, as carried out by individual cells.
signal transduction
The cascade of processes by which a signal interacts with a receptor, causing a change in the level or activity of a second messenger or other downstream target, and ultimately effecting a change in the functioning of the cell.
protein metabolic process
The chemical reactions and pathways involving a specific protein, rather than of proteins in general. Includes protein modification.
cellular biopolymer metabolic process
The chemical reactions and pathways involving biopolymers, long, repeating chains of monomers found in nature, such as polysaccharides and proteins, as carried out by individual cells.
cellular protein metabolic process
The chemical reactions and pathways involving a specific protein, rather than of proteins in general, occurring at the level of an individual cell. Includes protein modification.
protein modification process
The covalent alteration of one or more amino acids occurring in proteins, peptides and nascent polypeptides (co-translational, post-translational modifications). Includes the modification of charged tRNAs that are destined to occur in a protein (pre-translation modification).
positive regulation of transferase activity
Any process that activates or increases the frequency, rate or extent of transferase activity, the catalysis of the transfer of a group, e.g. a methyl group, glycosyl group, acyl group, phosphorus-containing, or other groups, from a donor compound to an acceptor.
positive regulation of kinase activity
Any process that activates or increases the frequency, rate or extent of kinase activity, the catalysis of the transfer of a phosphate group, usually from ATP, to a substrate molecule.
protein amino acid phosphorylation
The process of introducing a phosphate group on to a protein.
activation of MAPK activity
The initiation of the activity of the inactive enzyme MAP kinase by phosphorylation by a MAPKK.
positive regulation of protein kinase activity
Any process that activates or increases the frequency, rate or extent of protein kinase activity.
positive regulation of MAP kinase activity
Any process that activates or increases the frequency, rate or extent of MAP kinase activity.
activation of MAPK activity
The initiation of the activity of the inactive enzyme MAP kinase by phosphorylation by a MAPKK.

intracellular
The living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
cell
The basic structural and functional unit of all organisms. Includes the plasma membrane and any external encapsulating structures such as the cell wall and cell envelope.
nucleus
A membrane-bounded organelle of eukaryotic cells in which chromosomes are housed and replicated. In most cells, the nucleus contains all of the cell's chromosomes except the organellar chromosomes, and is the site of RNA synthesis and processing. In some species, or in specialized cell types, RNA metabolism or DNA replication may be absent.
cytoplasm
All of the contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures.
cytoskeleton
Any of the various filamentous elements that form the internal framework of cells, and typically remain after treatment of the cells with mild detergent to remove membrane constituents and soluble components of the cytoplasm. The term embraces intermediate filaments, microfilaments, microtubules, the microtrabecular lattice, and other structures characterized by a polymeric filamentous nature and long-range order within the cell. The various elements of the cytoskeleton not only serve in the maintenance of cellular shape but also have roles in other cellular functions, including cellular movement, cell division, endocytosis, and movement of organelles.
cell surface
The external part of the cell wall and/or plasma membrane.
cytoplasmic vesicle
A vesicle formed of membrane or protein, found in the cytoplasm of a cell.
vesicle
Any small, fluid-filled, spherical organelle enclosed by membrane or protein.
organelle
Organized structure of distinctive morphology and function. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton. Excludes the plasma membrane.
membrane-bounded organelle
Organized structure of distinctive morphology and function, bounded by a single or double lipid bilayer membrane. Includes the nucleus, mitochondria, plastids, vacuoles, and vesicles. Excludes the plasma membrane.
non-membrane-bounded organelle
Organized structure of distinctive morphology and function, not bounded by a lipid bilayer membrane. Includes ribosomes, the cytoskeleton and chromosomes.
intracellular organelle
Organized structure of distinctive morphology and function, occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton. Excludes the plasma membrane.
intracellular membrane-bounded organelle
Organized structure of distinctive morphology and function, bounded by a single or double lipid bilayer membrane and occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, and vesicles. Excludes the plasma membrane.
intracellular non-membrane-bounded organelle
Organized structure of distinctive morphology and function, not bounded by a lipid bilayer membrane and occurring within the cell. Includes ribosomes, the cytoskeleton and chromosomes.
organelle part
Any constituent part of an organelle, an organized structure of distinctive morphology and function. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton, but excludes the plasma membrane.
intracellular part
Any constituent part of the living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
nuclear part
Any constituent part of the nucleus, a membrane-bounded organelle of eukaryotic cells in which chromosomes are housed and replicated.
cytoplasmic part
Any constituent part of the cytoplasm, all of the contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures.
intracellular organelle part
A constituent part of an intracellular organelle, an organized structure of distinctive morphology and function, occurring within the cell. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton but excludes the plasma membrane.
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
all
This term is the most general term possible
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
organelle part
Any constituent part of an organelle, an organized structure of distinctive morphology and function. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton, but excludes the plasma membrane.
cytoplasmic vesicle
A vesicle formed of membrane or protein, found in the cytoplasm of a cell.
intracellular membrane-bounded organelle
Organized structure of distinctive morphology and function, bounded by a single or double lipid bilayer membrane and occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, and vesicles. Excludes the plasma membrane.
intracellular non-membrane-bounded organelle
Organized structure of distinctive morphology and function, not bounded by a lipid bilayer membrane and occurring within the cell. Includes ribosomes, the cytoskeleton and chromosomes.
intracellular organelle part
A constituent part of an intracellular organelle, an organized structure of distinctive morphology and function, occurring within the cell. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton but excludes the plasma membrane.
intracellular part
Any constituent part of the living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
intracellular organelle
Organized structure of distinctive morphology and function, occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton. Excludes the plasma membrane.
intracellular organelle part
A constituent part of an intracellular organelle, an organized structure of distinctive morphology and function, occurring within the cell. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton but excludes the plasma membrane.
cytoplasmic part
Any constituent part of the cytoplasm, all of the contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures.
cytoplasmic vesicle
A vesicle formed of membrane or protein, found in the cytoplasm of a cell.
nuclear part
Any constituent part of the nucleus, a membrane-bounded organelle of eukaryotic cells in which chromosomes are housed and replicated.

protein binding
Interacting selectively with any protein or protein complex (a complex of two or more proteins that may include other nonprotein molecules).
molecular_function
Elemental activities, such as catalysis or binding, describing the actions of a gene product at the molecular level. A given gene product may exhibit one or more molecular functions.
binding
The selective, often stoichiometric, interaction of a molecule with one or more specific sites on another molecule.
all
This term is the most general term possible
| Id | Pvalue | ExpCount | Count | Size |
|---|---|---|---|---|
| miR-320/320abcd | 2.028e-02 | 19.07 | 41 | 377 |
ABCB1ATP-binding cassette, sub-family B (MDR/TAP), member 1 (209993_at), score: 0.55 ACLYATP citrate lyase (210337_s_at), score: 0.6 ACTBactin, beta (AFFX-HSAC07/X00351_5_at), score: 0.49 ACTR2ARP2 actin-related protein 2 homolog (yeast) (200727_s_at), score: 0.71 ADAM10ADAM metallopeptidase domain 10 (214895_s_at), score: 0.66 ADAM15ADAM metallopeptidase domain 15 (217007_s_at), score: 0.57 ADRBK1adrenergic, beta, receptor kinase 1 (201401_s_at), score: 0.51 AFF4AF4/FMR2 family, member 4 (219199_at), score: 0.62 AGFG1ArfGAP with FG repeats 1 (213926_s_at), score: 0.66 AHCYL1S-adenosylhomocysteine hydrolase-like 1 (200848_at), score: 0.52 AKAP10A kinase (PRKA) anchor protein 10 (205045_at), score: 0.6 ANKFY1ankyrin repeat and FYVE domain containing 1 (219868_s_at), score: 0.66 API5apoptosis inhibitor 5 (214959_s_at), score: 0.51 ARF1ADP-ribosylation factor 1 (208750_s_at), score: 0.69 ARFGEF1ADP-ribosylation factor guanine nucleotide-exchange factor 1(brefeldin A-inhibited) (216266_s_at), score: 0.72 ARHGAP10Rho GTPase activating protein 10 (219431_at), score: -0.52 ARHGDIARho GDP dissociation inhibitor (GDI) alpha (213606_s_at), score: 0.57 ARHGEF11Rho guanine nucleotide exchange factor (GEF) 11 (202914_s_at), score: 0.48 ARL2BPADP-ribosylation factor-like 2 binding protein (202092_s_at), score: 0.49 ARNTLaryl hydrocarbon receptor nuclear translocator-like (209824_s_at), score: -0.55 ASAP1ArfGAP with SH3 domain, ankyrin repeat and PH domain 1 (221039_s_at), score: 0.54 ASPHaspartate beta-hydroxylase (205808_at), score: 0.7 ASXL1additional sex combs like 1 (Drosophila) (212237_at), score: 0.47 ATP13A3ATPase type 13A3 (219558_at), score: 0.75 ATP6AP2ATPase, H+ transporting, lysosomal accessory protein 2 (201444_s_at), score: 0.51 ATP6V1AATPase, H+ transporting, lysosomal 70kDa, V1 subunit A (201971_s_at), score: 0.83 ATP9AATPase, class II, type 9A (212062_at), score: -0.52 BCHEbutyrylcholinesterase (205433_at), score: 0.56 BCL2L1BCL2-like 1 (215037_s_at), score: 0.62 BCLAF1BCL2-associated transcription factor 1 (201101_s_at), score: 0.64 BET1Lblocked early in transport 1 homolog (S. cerevisiae)-like (220470_at), score: 0.53 BGNbiglycan (201262_s_at), score: 0.68 BICD2bicaudal D homolog 2 (Drosophila) (209203_s_at), score: 0.5 BMPR2bone morphogenetic protein receptor, type II (serine/threonine kinase) (210214_s_at), score: 0.48 BSGbasigin (Ok blood group) (208677_s_at), score: 0.54 BTRCbeta-transducin repeat containing (216091_s_at), score: 0.47 BZW1L1basic leucine zipper and W2 domains 1 like 1 (200776_s_at), score: 0.48 C10orf28chromosome 10 open reading frame 28 (210455_at), score: 0.53 C14orf118chromosome 14 open reading frame 118 (219720_s_at), score: 0.65 C18orf10chromosome 18 open reading frame 10 (213617_s_at), score: 0.62 C19orf6chromosome 19 open reading frame 6 (213986_s_at), score: 0.81 C20orf30chromosome 20 open reading frame 30 (220477_s_at), score: 0.6 CACNA2D1calcium channel, voltage-dependent, alpha 2/delta subunit 1 (207050_at), score: 0.67 CALM3calmodulin 3 (phosphorylase kinase, delta) (200622_x_at), score: 0.51 CALUcalumenin (214845_s_at), score: 0.69 CANXcalnexin (208853_s_at), score: 0.81 CAPRIN1cell cycle associated protein 1 (200722_s_at), score: 0.78 CBFBcore-binding factor, beta subunit (206788_s_at), score: 0.6 CCDC46coiled-coil domain containing 46 (213644_at), score: -0.51 CCDC6coiled-coil domain containing 6 (204716_at), score: 0.52 CCT2chaperonin containing TCP1, subunit 2 (beta) (201946_s_at), score: 0.51 CD164CD164 molecule, sialomucin (208653_s_at), score: 0.48 CD44CD44 molecule (Indian blood group) (210916_s_at), score: 0.76 CD46CD46 molecule, complement regulatory protein (211574_s_at), score: 0.74 CDC27cell division cycle 27 homolog (S. cerevisiae) (217881_s_at), score: 0.66 CDK6cyclin-dependent kinase 6 (207143_at), score: 0.56 CDV3CDV3 homolog (mouse) (213548_s_at), score: 0.88 CHMchoroideremia (Rab escort protein 1) (207099_s_at), score: 0.62 CLIC4chloride intracellular channel 4 (201559_s_at), score: 0.72 CLINT1clathrin interactor 1 (201768_s_at), score: 0.75 CLTCL1clathrin, heavy chain-like 1 (205944_s_at), score: -0.53 COL1A1collagen, type I, alpha 1 (217430_x_at), score: 0.54 COL6A1collagen, type VI, alpha 1 (212940_at), score: 0.76 COPAcoatomer protein complex, subunit alpha (214336_s_at), score: 0.91 COPS8COP9 constitutive photomorphogenic homolog subunit 8 (Arabidopsis) (202143_s_at), score: 0.52 CPDcarboxypeptidase D (201942_s_at), score: 0.87 CSNK1A1casein kinase 1, alpha 1 (206562_s_at), score: 0.69 CTTNcortactin (201059_at), score: -0.7 CYP51A1cytochrome P450, family 51, subfamily A, polypeptide 1 (216607_s_at), score: 0.71 CYTH3cytohesin 3 (206523_at), score: 0.52 DAB2disabled homolog 2, mitogen-responsive phosphoprotein (Drosophila) (201279_s_at), score: 0.48 DAPK3death-associated protein kinase 3 (203890_s_at), score: 0.64 DAZAP2DAZ associated protein 2 (212595_s_at), score: 0.78 DCBLD2discoidin, CUB and LCCL domain containing 2 (213865_at), score: 0.53 DCTN4dynactin 4 (p62) (218013_x_at), score: 0.63 DDX3XDEAD (Asp-Glu-Ala-Asp) box polypeptide 3, X-linked (201211_s_at), score: 1 DEGS1degenerative spermatocyte homolog 1, lipid desaturase (Drosophila) (207431_s_at), score: 0.54 DHDDSdehydrodolichyl diphosphate synthase (218547_at), score: -0.5 DHX9DEAH (Asp-Glu-Ala-His) box polypeptide 9 (212105_s_at), score: 0.94 DNAJB1DnaJ (Hsp40) homolog, subfamily B, member 1 (200664_s_at), score: 0.64 DND1dead end homolog 1 (zebrafish) (57739_at), score: 0.52 DSTdystonin (212253_x_at), score: 0.53 DTX3deltex homolog 3 (Drosophila) (49049_at), score: 0.48 DUSP10dual specificity phosphatase 10 (215501_s_at), score: 0.6 EDA2Rectodysplasin A2 receptor (221399_at), score: 0.48 EIF4Beukaryotic translation initiation factor 4B (219599_at), score: 0.48 EIF4G1eukaryotic translation initiation factor 4 gamma, 1 (208624_s_at), score: 0.52 ELK1ELK1, member of ETS oncogene family (210376_x_at), score: 0.49 ENSAendosulfine alpha (221487_s_at), score: 0.54 EPB41L2erythrocyte membrane protein band 4.1-like 2 (201718_s_at), score: 0.68 EPN2epsin 2 (203463_s_at), score: 0.48 EPRSglutamyl-prolyl-tRNA synthetase (200841_s_at), score: 0.62 ERLIN1ER lipid raft associated 1 (202444_s_at), score: 0.79 ETS1v-ets erythroblastosis virus E26 oncogene homolog 1 (avian) (214447_at), score: 0.54 EXOC5exocyst complex component 5 (218748_s_at), score: 0.67 EXTL3exostoses (multiple)-like 3 (211051_s_at), score: 0.48 EZRezrin (217234_s_at), score: 0.47 FAM108B1family with sequence similarity 108, member B1 (220285_at), score: 0.6 FBN1fibrillin 1 (202765_s_at), score: 0.56 FGFR1fibroblast growth factor receptor 1 (210973_s_at), score: 0.48 FHL1four and a half LIM domains 1 (201539_s_at), score: 0.49 FKBP15FK506 binding protein 15, 133kDa (76897_s_at), score: 0.66 FKBP1AFK506 binding protein 1A, 12kDa (210186_s_at), score: 0.54 FN1fibronectin 1 (214701_s_at), score: 0.82 FNTBfarnesyltransferase, CAAX box, beta (204764_at), score: 0.56 FRS2fibroblast growth factor receptor substrate 2 (221308_at), score: 0.61 FUSfusion (involved in t(12;16) in malignant liposarcoma) (217370_x_at), score: 0.63 FXR1fragile X mental retardation, autosomal homolog 1 (201635_s_at), score: 0.7 GABPAGA binding protein transcription factor, alpha subunit 60kDa (210188_at), score: 0.54 GDE1glycerophosphodiester phosphodiesterase 1 (202593_s_at), score: 0.64 GDI2GDP dissociation inhibitor 2 (200008_s_at), score: 0.71 GLE1GLE1 RNA export mediator homolog (yeast) (206920_s_at), score: 0.58 GLUD2glutamate dehydrogenase 2 (215794_x_at), score: 0.51 GLULglutamate-ammonia ligase (glutamine synthetase) (200648_s_at), score: 0.61 GNA12guanine nucleotide binding protein (G protein) alpha 12 (221737_at), score: -0.51 GNA13guanine nucleotide binding protein (G protein), alpha 13 (206917_at), score: 0.78 GNAI3guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 3 (201179_s_at), score: 0.48 GNB1guanine nucleotide binding protein (G protein), beta polypeptide 1 (200744_s_at), score: 0.52 GNSglucosamine (N-acetyl)-6-sulfatase (203676_at), score: 0.85 GOLGA3golgi autoantigen, golgin subfamily a, 3 (202106_at), score: -0.56 GPR137G protein-coupled receptor 137 (43934_at), score: 0.81 GPR176G protein-coupled receptor 176 (206673_at), score: 0.54 GTF2Igeneral transcription factor II, i (210892_s_at), score: 0.87 H2AFYH2A histone family, member Y (214500_at), score: 0.57 HADHAhydroxyacyl-Coenzyme A dehydrogenase/3-ketoacyl-Coenzyme A thiolase/enoyl-Coenzyme A hydratase (trifunctional protein), alpha subunit (208629_s_at), score: 0.58 HERC4hect domain and RLD 4 (208055_s_at), score: 0.53 HIPK1homeodomain interacting protein kinase 1 (212291_at), score: 0.48 HIPK3homeodomain interacting protein kinase 3 (210148_at), score: 0.61 HMGA1high mobility group AT-hook 1 (210457_x_at), score: 0.57 HNRNPCheterogeneous nuclear ribonucleoprotein C (C1/C2) (200751_s_at), score: 0.66 HNRNPH1heterogeneous nuclear ribonucleoprotein H1 (H) (213470_s_at), score: 0.59 HNRNPUheterogeneous nuclear ribonucleoprotein U (scaffold attachment factor A) (200593_s_at), score: 0.53 HP1BP3heterochromatin protein 1, binding protein 3 (220633_s_at), score: 0.79 HSPA4heat shock 70kDa protein 4 (211016_x_at), score: 0.8 HSPA8heat shock 70kDa protein 8 (210338_s_at), score: 0.5 HSPD1heat shock 60kDa protein 1 (chaperonin) (200806_s_at), score: 0.53 HTATSF1HIV-1 Tat specific factor 1 (202601_s_at), score: 0.48 IFI16interferon, gamma-inducible protein 16 (208965_s_at), score: 0.57 IFNGR1interferon gamma receptor 1 (211676_s_at), score: 0.56 IL13RA1interleukin 13 receptor, alpha 1 (210904_s_at), score: 0.51 IL6STinterleukin 6 signal transducer (gp130, oncostatin M receptor) (204864_s_at), score: 0.83 ILF3interleukin enhancer binding factor 3, 90kDa (208930_s_at), score: 0.47 IPO8importin 8 (205701_at), score: 0.65 IQGAP1IQ motif containing GTPase activating protein 1 (213446_s_at), score: 0.46 IREB2iron-responsive element binding protein 2 (214666_x_at), score: 0.47 KCTD20potassium channel tetramerisation domain containing 20 (214849_at), score: 0.52 KIAA0649KIAA0649 (203955_at), score: -0.54 KIAA1033KIAA1033 (215936_s_at), score: 0.61 KIDINS220kinase D-interacting substrate, 220kDa (212162_at), score: 0.56 KIFC3kinesin family member C3 (209661_at), score: -0.55 KITLGKIT ligand (211124_s_at), score: 0.69 KLHL20kelch-like 20 (Drosophila) (210635_s_at), score: 0.51 KPNA4karyopherin alpha 4 (importin alpha 3) (209653_at), score: 0.66 LIG4ligase IV, DNA, ATP-dependent (206235_at), score: 0.54 LMAN1lectin, mannose-binding, 1 (203294_s_at), score: 0.81 LOC100133166similar to neighbor of BRCA1 gene 1 (201383_s_at), score: 0.51 LOC100133724similar to Voltage-dependent anion-selective channel protein 1 (VDAC-1) (hVDAC1) (Outer mitochondrial membrane protein porin 1) (Plasmalemmal porin) (Porin 31HL) (Porin 31HM) (217140_s_at), score: 0.52 LOC285412similar to KIAA0737 protein (217448_s_at), score: 0.75 LOC440434hypothetical protein FLJ11822 (215090_x_at), score: 0.47 LOC727977hypothetical LOC727977 (205787_x_at), score: 0.58 LOC729148nuclear undecaprenyl pyrophosphate synthase 1 pseudogene (215207_x_at), score: 0.61 LOC730092RRN3 RNA polymerase I transcription factor homolog (S. cerevisiae) pseudogene (216908_x_at), score: 0.71 LOXlysyl oxidase (213640_s_at), score: 0.5 LOXL2lysyl oxidase-like 2 (202997_s_at), score: 0.53 LRP1low density lipoprotein-related protein 1 (alpha-2-macroglobulin receptor) (200784_s_at), score: 0.48 LRRC47leucine rich repeat containing 47 (212904_at), score: -0.51 LRRN2leucine rich repeat neuronal 2 (216164_at), score: 0.51 LSSlanosterol synthase (2,3-oxidosqualene-lanosterol cyclase) (211019_s_at), score: 0.58 M6PRmannose-6-phosphate receptor (cation dependent) (200900_s_at), score: 0.68 MACF1microtubule-actin crosslinking factor 1 (208633_s_at), score: 0.57 MADDMAP-kinase activating death domain (38398_at), score: -0.75 MAFFv-maf musculoaponeurotic fibrosarcoma oncogene homolog F (avian) (36711_at), score: -0.52 MAFGv-maf musculoaponeurotic fibrosarcoma oncogene homolog G (avian) (204970_s_at), score: -0.53 MAGOHmago-nashi homolog, proliferation-associated (Drosophila) (210092_at), score: 0.48 MAP3K2mitogen-activated protein kinase kinase kinase 2 (221695_s_at), score: 0.72 MAP3K7mitogen-activated protein kinase kinase kinase 7 (211537_x_at), score: 0.74 MAP3K7IP2mitogen-activated protein kinase kinase kinase 7 interacting protein 2 (210284_s_at), score: 0.83 MAP4K5mitogen-activated protein kinase kinase kinase kinase 5 (203553_s_at), score: 0.69 MAPK1mitogen-activated protein kinase 1 (208351_s_at), score: 0.61 MAPKAPK2mitogen-activated protein kinase-activated protein kinase 2 (201461_s_at), score: 0.68 MARCH7membrane-associated ring finger (C3HC4) 7 (202654_x_at), score: 0.68 MAT2Amethionine adenosyltransferase II, alpha (200769_s_at), score: 0.77 MAXMYC associated factor X (210734_x_at), score: 0.67 MAZMYC-associated zinc finger protein (purine-binding transcription factor) (207824_s_at), score: 0.62 MBNL1muscleblind-like (Drosophila) (201151_s_at), score: 0.66 MBNL2muscleblind-like 2 (Drosophila) (205018_s_at), score: 0.71 MBTPS2membrane-bound transcription factor peptidase, site 2 (206473_at), score: 0.65 ME2malic enzyme 2, NAD(+)-dependent, mitochondrial (210154_at), score: 0.48 MED6mediator complex subunit 6 (207079_s_at), score: 0.52 METmet proto-oncogene (hepatocyte growth factor receptor) (211599_x_at), score: 0.54 MFN2mitofusin 2 (216205_s_at), score: 0.72 MOBKL1BMOB1, Mps One Binder kinase activator-like 1B (yeast) (201299_s_at), score: 0.63 MPDU1mannose-P-dolichol utilization defect 1 (209208_at), score: 0.47 MT1Mmetallothionein 1M (217546_at), score: -0.58 MTDHmetadherin (212251_at), score: -0.51 MTMR1myotubularin related protein 1 (214975_s_at), score: 0.54 MYO5Bmyosin VB (202002_at), score: 0.47 NAB1NGFI-A binding protein 1 (EGR1 binding protein 1) (208047_s_at), score: 0.55 NBPF10neuroblastoma breakpoint family, member 10 (214693_x_at), score: 0.57 NBPF14neuroblastoma breakpoint family, member 14 (201104_x_at), score: 0.55 NCOA2nuclear receptor coactivator 2 (205732_s_at), score: 0.65 NCRNA00084non-protein coding RNA 84 (214657_s_at), score: 0.46 NF1neurofibromin 1 (211094_s_at), score: 0.63 NID1nidogen 1 (202008_s_at), score: 0.51 NNTnicotinamide nucleotide transhydrogenase (202784_s_at), score: 0.59 NONOnon-POU domain containing, octamer-binding (208698_s_at), score: 0.7 NOTCH2Notch homolog 2 (Drosophila) (210756_s_at), score: 0.85 NRASneuroblastoma RAS viral (v-ras) oncogene homolog (202647_s_at), score: 0.48 NUMA1nuclear mitotic apparatus protein 1 (214251_s_at), score: 0.65 OASL2'-5'-oligoadenylate synthetase-like (210797_s_at), score: 0.52 OSMRoncostatin M receptor (205729_at), score: 0.7 PAFAH1B1platelet-activating factor acetylhydrolase, isoform Ib, alpha subunit 45kDa (211547_s_at), score: 0.81 PALLDpalladin, cytoskeletal associated protein (200906_s_at), score: 0.47 PAPOLApoly(A) polymerase alpha (212720_at), score: 0.7 PCDHGA11protocadherin gamma subfamily A, 11 (211876_x_at), score: 0.89 PCDHGA3protocadherin gamma subfamily A, 3 (216352_x_at), score: 0.89 PCIF1PDX1 C-terminal inhibiting factor 1 (222045_s_at), score: 0.54 PDAP1PDGFA associated protein 1 (202290_at), score: 0.62 PDLIM4PDZ and LIM domain 4 (214175_x_at), score: -0.52 PDPK13-phosphoinositide dependent protein kinase-1 (204524_at), score: -0.59 PEA15phosphoprotein enriched in astrocytes 15 (200787_s_at), score: 0.56 PGRMC1progesterone receptor membrane component 1 (201120_s_at), score: 0.62 PHF17PHD finger protein 17 (218517_at), score: -0.51 PHTF2putative homeodomain transcription factor 2 (217097_s_at), score: 0.84 PICALMphosphatidylinositol binding clathrin assembly protein (215236_s_at), score: 0.74 POLDIP3polymerase (DNA-directed), delta interacting protein 3 (215357_s_at), score: 0.65 POM121POM121 membrane glycoprotein (rat) (212178_s_at), score: 0.55 PON2paraoxonase 2 (210830_s_at), score: 0.54 PPP2R5Dprotein phosphatase 2, regulatory subunit B', delta isoform (211159_s_at), score: 0.51 PRIM2primase, DNA, polypeptide 2 (58kDa) (215708_s_at), score: 0.47 PRKAA1protein kinase, AMP-activated, alpha 1 catalytic subunit (209799_at), score: 0.57 PRKAB1protein kinase, AMP-activated, beta 1 non-catalytic subunit (201835_s_at), score: 0.63 PRKAR1Aprotein kinase, cAMP-dependent, regulatory, type I, alpha (tissue specific extinguisher 1) (200604_s_at), score: 0.69 PRKD3protein kinase D3 (211084_x_at), score: 0.74 PRPF4PRP4 pre-mRNA processing factor 4 homolog (yeast) (209162_s_at), score: 0.66 PRPF40APRP40 pre-mRNA processing factor 40 homolog A (S. cerevisiae) (214941_s_at), score: 0.58 PRPF4BPRP4 pre-mRNA processing factor 4 homolog B (yeast) (211090_s_at), score: 0.63 PRPF6PRP6 pre-mRNA processing factor 6 homolog (S. cerevisiae) (208879_x_at), score: 0.52 PRRX1paired related homeobox 1 (205991_s_at), score: 0.7 PSAPprosaposin (200866_s_at), score: 0.47 PSEN1presenilin 1 (207782_s_at), score: 0.62 PSME4proteasome (prosome, macropain) activator subunit 4 (212220_at), score: 0.53 PTBP1polypyrimidine tract binding protein 1 (212016_s_at), score: 0.87 PTP4A1protein tyrosine phosphatase type IVA, member 1 (200730_s_at), score: 0.61 PTPLAD1protein tyrosine phosphatase-like A domain containing 1 (217777_s_at), score: 0.61 PTPN11protein tyrosine phosphatase, non-receptor type 11 (209896_s_at), score: 0.56 PTPN12protein tyrosine phosphatase, non-receptor type 12 (216915_s_at), score: 0.59 PTPRAprotein tyrosine phosphatase, receptor type, A (213799_s_at), score: 0.57 PTPROprotein tyrosine phosphatase, receptor type, O (211600_at), score: 0.59 PXNpaxillin (211823_s_at), score: 0.71 RAB11BRAB11B, member RAS oncogene family (34478_at), score: 0.48 RAB4ARAB4A, member RAS oncogene family (203582_s_at), score: 0.5 RAB5ARAB5A, member RAS oncogene family (206113_s_at), score: 0.89 RABGGTBRab geranylgeranyltransferase, beta subunit (213704_at), score: -0.61 RALBv-ral simian leukemia viral oncogene homolog B (ras related; GTP binding protein) (202101_s_at), score: 0.46 RANBP2RAN binding protein 2 (201711_x_at), score: 0.57 RASA3RAS p21 protein activator 3 (206220_s_at), score: 0.58 RBM9RNA binding motif protein 9 (213901_x_at), score: 0.49 RCC1regulator of chromosome condensation 1 (206499_s_at), score: 0.46 RCOR3REST corepressor 3 (218344_s_at), score: 0.5 RDXradixin (204969_s_at), score: 0.55 RFX7regulatory factor X, 7 (218430_s_at), score: 0.47 RIN2Ras and Rab interactor 2 (209684_at), score: -0.54 RNF146ring finger protein 146 (221430_s_at), score: 0.51 RNF19Aring finger protein 19A (220483_s_at), score: 0.62 RNFT1ring finger protein, transmembrane 1 (221194_s_at), score: 0.6 RNGTTRNA guanylyltransferase and 5'-phosphatase (204207_s_at), score: 0.5 RRN3RRN3 RNA polymerase I transcription factor homolog (S. cerevisiae) (216902_s_at), score: 0.64 RRP7Bribosomal RNA processing 7 homolog B (S. cerevisiae) (214828_s_at), score: 0.59 RXRBretinoid X receptor, beta (215099_s_at), score: 0.74 RYKRYK receptor-like tyrosine kinase (216976_s_at), score: 0.5 SART3squamous cell carcinoma antigen recognized by T cells 3 (209127_s_at), score: 0.52 SCAMP1secretory carrier membrane protein 1 (206667_s_at), score: 0.82 SCARB2scavenger receptor class B, member 2 (201647_s_at), score: 0.77 SCYL2SCY1-like 2 (S. cerevisiae) (221220_s_at), score: 0.63 SDHCsuccinate dehydrogenase complex, subunit C, integral membrane protein, 15kDa (216591_s_at), score: 0.72 SEC23ASec23 homolog A (S. cerevisiae) (204344_s_at), score: 0.54 SEC23IPSEC23 interacting protein (216392_s_at), score: 0.57 SELTselenoprotein T (217811_at), score: 0.47 SENP3SUMO1/sentrin/SMT3 specific peptidase 3 (215113_s_at), score: 0.61 SEPT10septin 10 (214720_x_at), score: 0.62 SEPT11septin 11 (201308_s_at), score: 0.72 SEPT2septin 2 (200778_s_at), score: 0.67 SETD3SET domain containing 3 (212465_at), score: -0.53 SFRS1splicing factor, arginine/serine-rich 1 (201742_x_at), score: 0.75 SFRS5splicing factor, arginine/serine-rich 5 (212266_s_at), score: 0.54 SGCBsarcoglycan, beta (43kDa dystrophin-associated glycoprotein) (205121_at), score: 0.62 SGPP1sphingosine-1-phosphate phosphatase 1 (221268_s_at), score: 0.55 SHC1SHC (Src homology 2 domain containing) transforming protein 1 (201469_s_at), score: 0.53 SLC12A4solute carrier family 12 (potassium/chloride transporters), member 4 (209401_s_at), score: 0.61 SLC2A1solute carrier family 2 (facilitated glucose transporter), member 1 (201250_s_at), score: -0.5 SLC39A6solute carrier family 39 (zinc transporter), member 6 (202089_s_at), score: 0.5 SLC39A9solute carrier family 39 (zinc transporter), member 9 (217859_s_at), score: 0.47 SMAD5SMAD family member 5 (205187_at), score: 0.59 SMARCA2SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 (212257_s_at), score: 0.51 SMC3structural maintenance of chromosomes 3 (209257_s_at), score: 0.5 SMEK1SMEK homolog 1, suppressor of mek1 (Dictyostelium) (220368_s_at), score: 0.92 SMG1SMG1 homolog, phosphatidylinositol 3-kinase-related kinase (C. elegans) (210057_at), score: 0.51 SNAP23synaptosomal-associated protein, 23kDa (214544_s_at), score: 0.77 SOCS5suppressor of cytokine signaling 5 (209647_s_at), score: 0.54 SP1Sp1 transcription factor (214732_at), score: 0.49 SPAG9sperm associated antigen 9 (206748_s_at), score: 0.68 SPPL2Bsignal peptide peptidase-like 2B (215833_s_at), score: 0.6 SPSB3splA/ryanodine receptor domain and SOCS box containing 3 (46256_at), score: -0.57 SPTBN1spectrin, beta, non-erythrocytic 1 (200672_x_at), score: 0.61 SPTLC1serine palmitoyltransferase, long chain base subunit 1 (202278_s_at), score: 0.63 SRBD1S1 RNA binding domain 1 (219055_at), score: -0.53 SRPRsignal recognition particle receptor (docking protein) (200917_s_at), score: 0.69 SS18synovial sarcoma translocation, chromosome 18 (216684_s_at), score: 0.81 STAT1signal transducer and activator of transcription 1, 91kDa (AFFX-HUMISGF3A/M97935_MA_at), score: 0.54 STIP1stress-induced-phosphoprotein 1 (212009_s_at), score: 0.86 TAF12TAF12 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 20kDa (209463_s_at), score: 0.47 TAF9BTAF9B RNA polymerase II, TATA box binding protein (TBP)-associated factor, 31kDa (221618_s_at), score: 0.68 TBL1XR1transducin (beta)-like 1 X-linked receptor 1 (221428_s_at), score: 0.61 TERF2IPtelomeric repeat binding factor 2, interacting protein (201174_s_at), score: -0.55 TFCP2transcription factor CP2 (207627_s_at), score: 0.46 TGFBR2transforming growth factor, beta receptor II (70/80kDa) (207334_s_at), score: 0.82 TJP1tight junction protein 1 (zona occludens 1) (214168_s_at), score: 0.62 TLN1talin 1 (203254_s_at), score: 0.48 TM9SF4transmembrane 9 superfamily protein member 4 (212194_s_at), score: 0.59 TMCC1transmembrane and coiled-coil domain family 1 (213351_s_at), score: -0.51 TMED2transmembrane emp24 domain trafficking protein 2 (204426_at), score: 0.73 TMEM97transmembrane protein 97 (212279_at), score: 0.5 TMOD3tropomodulin 3 (ubiquitous) (220800_s_at), score: 0.66 TMPOthymopoietin (209754_s_at), score: 0.47 TMX1thioredoxin-related transmembrane protein 1 (208097_s_at), score: 0.66 TNFAIP1tumor necrosis factor, alpha-induced protein 1 (endothelial) (201208_s_at), score: 0.5 TOR1AIP1torsin A interacting protein 1 (216100_s_at), score: 0.59 TPM4tropomyosin 4 (212481_s_at), score: 0.61 TRAM1translocation associated membrane protein 1 (210733_at), score: 0.55 TRIM25tripartite motif-containing 25 (206911_at), score: 0.5 TSNtranslin (201504_s_at), score: 0.63 TSPAN3tetraspanin 3 (200973_s_at), score: 0.47 TUBB2Atubulin, beta 2A (209372_x_at), score: 0.56 TWF1twinfilin, actin-binding protein, homolog 1 (Drosophila) (214007_s_at), score: 0.71 TXNRD3thioredoxin reductase 3 (59631_at), score: -0.56 UBAP2Lubiquitin associated protein 2-like (201378_s_at), score: 0.57 UBE2Aubiquitin-conjugating enzyme E2A (RAD6 homolog) (201898_s_at), score: 0.46 UBE2D3ubiquitin-conjugating enzyme E2D 3 (UBC4/5 homolog, yeast) (200669_s_at), score: 0.81 UBE2Hubiquitin-conjugating enzyme E2H (UBC8 homolog, yeast) (221962_s_at), score: 0.62 UBXN4UBX domain protein 4 (212008_at), score: 0.79 USP34ubiquitin specific peptidase 34 (212065_s_at), score: 0.6 UTRNutrophin (213022_s_at), score: 0.67 VAMP3vesicle-associated membrane protein 3 (cellubrevin) (201337_s_at), score: 0.81 WACWW domain containing adaptor with coiled-coil (219679_s_at), score: 0.9 WDR1WD repeat domain 1 (210935_s_at), score: 0.7 XPNPEP1X-prolyl aminopeptidase (aminopeptidase P) 1, soluble (208453_s_at), score: 0.52 YIPF5Yip1 domain family, member 5 (221423_s_at), score: 0.53 YWHAZtyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide (200641_s_at), score: 0.76 ZFAND5zinc finger, AN1-type domain 5 (217741_s_at), score: 0.79 ZFP36L2zinc finger protein 36, C3H type-like 2 (201367_s_at), score: 0.49 ZFRzinc finger RNA binding protein (33148_at), score: 0.53 ZMIZ2zinc finger, MIZ-type containing 2 (54970_at), score: -0.65 ZMYND11zinc finger, MYND domain containing 11 (202137_s_at), score: 0.75 ZNF148zinc finger protein 148 (203319_s_at), score: 0.48 ZNF224zinc finger protein 224 (216983_s_at), score: 0.51 ZNF26zinc finger protein 26 (219595_at), score: -0.53 ZNF629zinc finger protein 629 (213196_at), score: -0.62 ZNF675zinc finger protein 675 (217547_x_at), score: 0.52
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| 5042_CNTL.CEL | 6 | 8 | WBS | hgu133plus2 | none | WBS 1 |
| ctrl c 08-03.CEL | 3 | 1 | DS-CC | hgu133a | none | DS-CC 3 |
| E-TABM-263-raw-cel-1515486031.cel | 20 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| 4Twin.CEL | 4 | 2 | DS-twin | hgu133plus2 | none | DS-twin 4 |
| E-GEOD-3407-raw-cel-1437949938.cel | 8 | 4 | Cockayne | hgu133a | none | CSB |
| E-GEOD-3407-raw-cel-1437949854.cel | 7 | 4 | Cockayne | hgu133a | CS | eGFP |