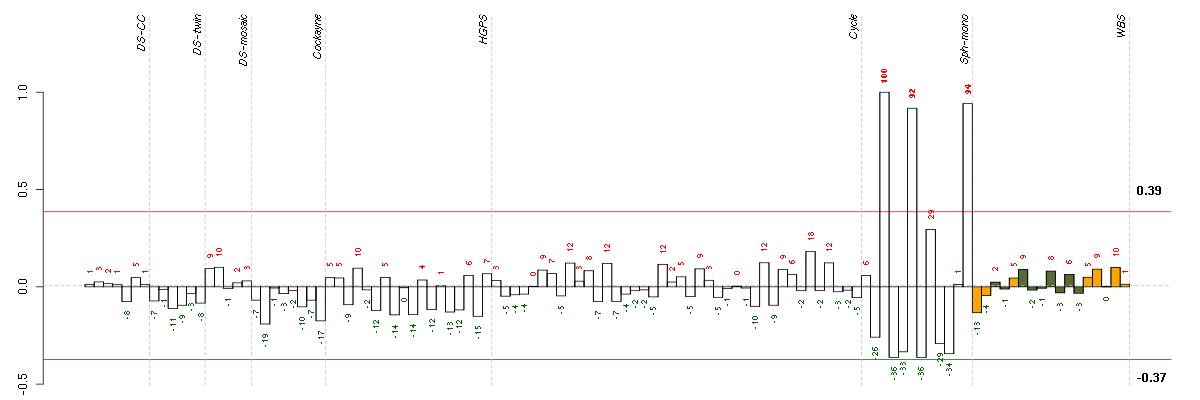

Under-expression is coded with green,
over-expression with red color.

immune system process
Any process involved in the development or functioning of the immune system, an organismal system for calibrated responses to potential internal or invasive threats.
antigen processing and presentation of peptide antigen via MHC class I
The process by which an antigen-presenting cell expresses a peptide antigen on its cell surface in association with an MHC class I protein complex. Class I here refers to classical class I molecules.
biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
antigen processing and presentation
The process by which an antigen-presenting cell expresses antigen (peptide or lipid) on its cell surface in association with an MHC protein complex.
antigen processing and presentation of peptide antigen
The process by which an antigen-presenting cell expresses peptide antigen in association with an MHC protein complex on its cell surface, including proteolysis and transport steps for the peptide antigen both prior to and following assembly with the MHC protein complex. The peptide antigen is typically, but not always, processed from an endogenous or exogenous protein.
all
This term is the most general term possible

plasma membrane
The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins.
membrane
Double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
MHC class I protein complex
A transmembrane protein complex composed of a MHC class I alpha chain and an invariant beta2-microglobin chain, and with or without a bound peptide antigen. Class I here refers to classical class I molecules.
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
cell
The basic structural and functional unit of all organisms. Includes the plasma membrane and any external encapsulating structures such as the cell wall and cell envelope.
macromolecular complex
A stable assembly of two or more macromolecules, i.e. proteins, nucleic acids, carbohydrates or lipids, in which the constituent parts function together.
MHC protein complex
A transmembrane protein complex composed of an MHC alpha chain and, in most cases, either an MHC class II beta chain or an invariant beta2-microglobin chain, and with or without a bound peptide, lipid, or polysaccharide antigen.
protein complex
Any macromolecular complex composed of two or more polypeptide subunits, which may or may not be identical. Protein complexes may have other associated non-protein prosthetic groups, such as nucleotides, metal ions or carbohydrate groups.
membrane part
Any constituent part of a membrane, a double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
plasma membrane part
Any constituent part of the plasma membrane, the membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins.
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
all
This term is the most general term possible
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
membrane part
Any constituent part of a membrane, a double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
plasma membrane part
Any constituent part of the plasma membrane, the membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins.
MHC protein complex
A transmembrane protein complex composed of an MHC alpha chain and, in most cases, either an MHC class II beta chain or an invariant beta2-microglobin chain, and with or without a bound peptide, lipid, or polysaccharide antigen.

molecular_function
Elemental activities, such as catalysis or binding, describing the actions of a gene product at the molecular level. A given gene product may exhibit one or more molecular functions.
signal transducer activity
Mediates the transfer of a signal from the outside to the inside of a cell by means other than the introduction of the signal molecule itself into the cell.
receptor activity
Combining with an extracellular or intracellular messenger to initiate a change in cell activity.
transmembrane receptor activity
Combining with an extracellular or intracellular messenger to initiate a change in cell activity, and spanning to the membrane of either the cell or an organelle.
MHC class I receptor activity
Combining with an MHC class I protein complex to initiate a change in cellular activity. Class I here refers to classical class I molecules.
molecular transducer activity
The molecular function that accepts an input of one form and creates an output of a different form.
all
This term is the most general term possible
| Id | Pvalue | ExpCount | Count | Size | Term |
|---|---|---|---|---|---|
| 04940 | 2.261e-02 | 0.6295 | 5 | 26 | Type I diabetes mellitus |
| 04612 | 3.804e-02 | 1.09 | 6 | 45 | Antigen processing and presentation |
ABAT4-aminobutyrate aminotransferase (209459_s_at), score: 0.62 ABCC10ATP-binding cassette, sub-family C (CFTR/MRP), member 10 (213485_s_at), score: 0.56 ABLIM1actin binding LIM protein 1 (200965_s_at), score: 0.75 ACOX3acyl-Coenzyme A oxidase 3, pristanoyl (204241_at), score: 0.55 ACTR8ARP8 actin-related protein 8 homolog (yeast) (218658_s_at), score: 0.59 ADAM9ADAM metallopeptidase domain 9 (meltrin gamma) (202381_at), score: 0.55 ADAMTS3ADAM metallopeptidase with thrombospondin type 1 motif, 3 (214913_at), score: 0.71 ADIPOR1adiponectin receptor 1 (217748_at), score: -0.6 AKAP12A kinase (PRKA) anchor protein 12 (210517_s_at), score: 0.54 ALCAMactivated leukocyte cell adhesion molecule (201951_at), score: 0.55 ALDH1A1aldehyde dehydrogenase 1 family, member A1 (212224_at), score: 0.86 ANXA3annexin A3 (209369_at), score: 0.57 APAF1apoptotic peptidase activating factor 1 (204859_s_at), score: 0.58 ARL15ADP-ribosylation factor-like 15 (219842_at), score: 0.81 ARMCX1armadillo repeat containing, X-linked 1 (218694_at), score: 0.57 ARSJarylsulfatase family, member J (219973_at), score: 0.6 ASAP2ArfGAP with SH3 domain, ankyrin repeat and PH domain 2 (206414_s_at), score: 0.55 ASCC3activating signal cointegrator 1 complex subunit 3 (212815_at), score: 0.58 ASPNasporin (219087_at), score: 0.6 ATF7IP2activating transcription factor 7 interacting protein 2 (219870_at), score: 0.57 ATG2BATG2 autophagy related 2 homolog B (S. cerevisiae) (219164_s_at), score: 0.55 ATP2B4ATPase, Ca++ transporting, plasma membrane 4 (212135_s_at), score: 0.56 ATP6V0BATPase, H+ transporting, lysosomal 21kDa, V0 subunit b (200078_s_at), score: -0.5 B2Mbeta-2-microglobulin (216231_s_at), score: -0.51 BAZ2Bbromodomain adjacent to zinc finger domain, 2B (203080_s_at), score: 0.54 BEX1brain expressed, X-linked 1 (218332_at), score: 0.55 BHLHB9basic helix-loop-helix domain containing, class B, 9 (213709_at), score: 0.54 BTN3A1butyrophilin, subfamily 3, member A1 (209770_at), score: 0.54 C10orf116chromosome 10 open reading frame 116 (203571_s_at), score: 0.55 C14orf169chromosome 14 open reading frame 169 (219526_at), score: 0.69 C15orf29chromosome 15 open reading frame 29 (218791_s_at), score: 0.67 C20orf111chromosome 20 open reading frame 111 (209020_at), score: -0.57 C21orf7chromosome 21 open reading frame 7 (221211_s_at), score: 0.54 C2orf37chromosome 2 open reading frame 37 (220172_at), score: 0.64 C6orf211chromosome 6 open reading frame 211 (218195_at), score: 0.61 C7orf23chromosome 7 open reading frame 23 (204215_at), score: -0.51 C9orf125chromosome 9 open reading frame 125 (213386_at), score: 0.56 CAND1cullin-associated and neddylation-dissociated 1 (208839_s_at), score: 0.64 CAND2cullin-associated and neddylation-dissociated 2 (putative) (213547_at), score: 0.72 CARD8caspase recruitment domain family, member 8 (204950_at), score: 0.58 CBFA2T2core-binding factor, runt domain, alpha subunit 2; translocated to, 2 (207625_s_at), score: -0.54 CD44CD44 molecule (Indian blood group) (210916_s_at), score: -0.59 CELSR3cadherin, EGF LAG seven-pass G-type receptor 3 (flamingo homolog, Drosophila) (40020_at), score: -0.63 CHRDL1chordin-like 1 (209763_at), score: 0.67 CLASP2cytoplasmic linker associated protein 2 (212306_at), score: 0.63 CLEC2BC-type lectin domain family 2, member B (209732_at), score: -0.59 COBLL1COBL-like 1 (203642_s_at), score: 0.8 CORO1Ccoronin, actin binding protein, 1C (221676_s_at), score: 0.55 CPEcarboxypeptidase E (201117_s_at), score: -0.55 CRIM1cysteine rich transmembrane BMP regulator 1 (chordin-like) (202552_s_at), score: 0.58 CRLF3cytokine receptor-like factor 3 (205474_at), score: 0.54 CTNND1catenin (cadherin-associated protein), delta 1 (208407_s_at), score: 0.55 CTSScathepsin S (202901_x_at), score: -0.57 CUGBP2CUG triplet repeat, RNA binding protein 2 (202158_s_at), score: 0.57 DAPK1death-associated protein kinase 1 (203139_at), score: 0.7 DDAH1dimethylarginine dimethylaminohydrolase 1 (209094_at), score: 0.66 DGKDdiacylglycerol kinase, delta 130kDa (208072_s_at), score: -0.61 DLX2distal-less homeobox 2 (207147_at), score: -0.51 DNAJC15DnaJ (Hsp40) homolog, subfamily C, member 15 (218435_at), score: 0.57 DNAJC6DnaJ (Hsp40) homolog, subfamily C, member 6 (204720_s_at), score: 0.66 DNASE1L1deoxyribonuclease I-like 1 (203912_s_at), score: 0.56 DRAMdamage-regulated autophagy modulator (218627_at), score: 0.55 DRG1developmentally regulated GTP binding protein 1 (202810_at), score: -0.51 EFHD1EF-hand domain family, member D1 (209343_at), score: 0.58 EIF1eukaryotic translation initiation factor 1 (212130_x_at), score: -0.54 EIF3Deukaryotic translation initiation factor 3, subunit D (200005_at), score: -0.56 ENOSF1enolase superfamily member 1 (204142_at), score: 0.71 EPHX1epoxide hydrolase 1, microsomal (xenobiotic) (202017_at), score: -0.53 ERGIC3ERGIC and golgi 3 (216032_s_at), score: -0.54 EXOGendo/exonuclease (5'-3'), endonuclease G-like (205521_at), score: 0.58 FABP3fatty acid binding protein 3, muscle and heart (mammary-derived growth inhibitor) (214285_at), score: 0.6 FAF1Fas (TNFRSF6) associated factor 1 (218080_x_at), score: 0.56 FAM105Afamily with sequence similarity 105, member A (219694_at), score: -0.51 FAM174Bfamily with sequence similarity 174, member B (51158_at), score: 1 FAM46Cfamily with sequence similarity 46, member C (220306_at), score: 0.89 FAM65Bfamily with sequence similarity 65, member B (209829_at), score: 0.84 FBN2fibrillin 2 (203184_at), score: 0.6 FLT1fms-related tyrosine kinase 1 (vascular endothelial growth factor/vascular permeability factor receptor) (222033_s_at), score: 0.79 FOXG1forkhead box G1 (206018_at), score: -0.51 FOXL2forkhead box L2 (220102_at), score: -0.58 FYCO1FYVE and coiled-coil domain containing 1 (218204_s_at), score: 0.56 FZD4frizzled homolog 4 (Drosophila) (218665_at), score: 0.56 GAL3ST4galactose-3-O-sulfotransferase 4 (219815_at), score: 0.74 GALCgalactosylceramidase (204417_at), score: 0.54 GALNT3UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 3 (GalNAc-T3) (203397_s_at), score: 0.86 GFRA1GDNF family receptor alpha 1 (205696_s_at), score: 0.83 GIPC2GIPC PDZ domain containing family, member 2 (219970_at), score: 0.67 GLT25D2glycosyltransferase 25 domain containing 2 (209883_at), score: 0.61 GNPTABN-acetylglucosamine-1-phosphate transferase, alpha and beta subunits (212959_s_at), score: 0.58 GPR126G protein-coupled receptor 126 (213094_at), score: 0.77 GRAMD3GRAM domain containing 3 (218706_s_at), score: 0.58 GREM1gremlin 1, cysteine knot superfamily, homolog (Xenopus laevis) (218469_at), score: 0.83 GRIK2glutamate receptor, ionotropic, kainate 2 (213845_at), score: 0.56 HEY1hairy/enhancer-of-split related with YRPW motif 1 (44783_s_at), score: -0.51 HLA-Bmajor histocompatibility complex, class I, B (211911_x_at), score: -0.6 HLA-Cmajor histocompatibility complex, class I, C (211799_x_at), score: -0.52 HLA-Emajor histocompatibility complex, class I, E (200904_at), score: -0.51 HLA-Fmajor histocompatibility complex, class I, F (204806_x_at), score: -0.52 HSD17B12hydroxysteroid (17-beta) dehydrogenase 12 (217869_at), score: 0.65 HYPKHuntingtin interacting protein K (218680_x_at), score: -0.52 IFIT5interferon-induced protein with tetratricopeptide repeats 5 (203596_s_at), score: 0.54 IGF2insulin-like growth factor 2 (somatomedin A) (202409_at), score: 0.54 IGSF3immunoglobulin superfamily, member 3 (202421_at), score: -0.56 IL23Ainterleukin 23, alpha subunit p19 (211796_s_at), score: 0.89 INAinternexin neuronal intermediate filament protein, alpha (204465_s_at), score: 0.71 INHBEinhibin, beta E (210587_at), score: 0.59 INPP4Binositol polyphosphate-4-phosphatase, type II, 105kDa (205376_at), score: 0.62 IPWimprinted in Prader-Willi syndrome (non-protein coding) (221974_at), score: 0.6 ISL1ISL LIM homeobox 1 (206104_at), score: 0.72 JMJD1Ajumonji domain containing 1A (212689_s_at), score: -0.53 KCNJ2potassium inwardly-rectifying channel, subfamily J, member 2 (206765_at), score: 0.75 KCNN2potassium intermediate/small conductance calcium-activated channel, subfamily N, member 2 (220116_at), score: 0.71 KIAA0802KIAA0802 (213358_at), score: 0.57 KIF3Bkinesin family member 3B (203943_at), score: -0.58 KLHL3kelch-like 3 (Drosophila) (221221_s_at), score: 0.63 KYNUkynureninase (L-kynurenine hydrolase) (217388_s_at), score: -0.73 L1CAML1 cell adhesion molecule (204584_at), score: 0.8 LEF1lymphoid enhancer-binding factor 1 (221558_s_at), score: -0.53 LMO7LIM domain 7 (202674_s_at), score: 0.72 LPPR4plasticity related gene 1 (213496_at), score: 0.76 LRRC1leucine rich repeat containing 1 (218816_at), score: 0.74 LRRC16Aleucine rich repeat containing 16A (219573_at), score: 0.68 LUMlumican (201744_s_at), score: 0.66 LXNlatexin (218729_at), score: 0.63 MAMLD1mastermind-like domain containing 1 (205088_at), score: 0.58 MAN1A2mannosidase, alpha, class 1A, member 2 (217921_at), score: 0.6 MANSC1MANSC domain containing 1 (220945_x_at), score: 0.81 MARCH3membrane-associated ring finger (C3HC4) 3 (213256_at), score: 0.54 ME1malic enzyme 1, NADP(+)-dependent, cytosolic (204058_at), score: 0.68 MED23mediator complex subunit 23 (218846_at), score: 0.55 MKRN2makorin ring finger protein 2 (218071_s_at), score: 0.59 MLF1myeloid leukemia factor 1 (204784_s_at), score: -0.58 MLLT3myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 3 (204918_s_at), score: 0.78 MOXD1monooxygenase, DBH-like 1 (209708_at), score: 0.56 MPPED2metallophosphoesterase domain containing 2 (205413_at), score: -0.51 MRPL52mitochondrial ribosomal protein L52 (221997_s_at), score: 0.63 MRPS18Cmitochondrial ribosomal protein S18C (220103_s_at), score: 0.71 MYO1Bmyosin IB (212364_at), score: 0.66 NACC2NACC family member 2, BEN and BTB (POZ) domain containing (212993_at), score: 0.55 NAV3neuron navigator 3 (204823_at), score: 0.59 NBEAneurobeachin (221207_s_at), score: 0.69 NBPF10neuroblastoma breakpoint family, member 10 (214693_x_at), score: -0.54 NDRG1N-myc downstream regulated 1 (200632_s_at), score: -0.53 NESnestin (218678_at), score: 0.71 NETO2neuropilin (NRP) and tolloid (TLL)-like 2 (218888_s_at), score: 0.55 NFE2L2nuclear factor (erythroid-derived 2)-like 2 (201146_at), score: -0.51 NLGN1neuroligin 1 (205893_at), score: 0.68 NPLOC4nuclear protein localization 4 homolog (S. cerevisiae) (217796_s_at), score: -0.51 NRN1neuritin 1 (218625_at), score: -0.61 NUAK2NUAK family, SNF1-like kinase, 2 (220987_s_at), score: 0.58 NUDT6nudix (nucleoside diphosphate linked moiety X)-type motif 6 (220183_s_at), score: 0.56 OXTRoxytocin receptor (206825_at), score: 0.59 PARGpoly (ADP-ribose) glycohydrolase (205060_at), score: 0.54 PBXIP1pre-B-cell leukemia homeobox interacting protein 1 (214176_s_at), score: 0.68 PCGF1polycomb group ring finger 1 (210023_s_at), score: -0.51 PDGFRAplatelet-derived growth factor receptor, alpha polypeptide (203131_at), score: 0.55 PEX5peroxisomal biogenesis factor 5 (203244_at), score: -0.6 PFDN2prefoldin subunit 2 (218336_at), score: -0.57 PGAP1post-GPI attachment to proteins 1 (213469_at), score: 0.62 PIGNphosphatidylinositol glycan anchor biosynthesis, class N (219048_at), score: 0.55 PLCB1phospholipase C, beta 1 (phosphoinositide-specific) (213222_at), score: 0.68 PLP1proteolipid protein 1 (210198_s_at), score: 0.55 POM121CPOM121 membrane glycoprotein C (213360_s_at), score: -0.51 PPARGC1Aperoxisome proliferator-activated receptor gamma, coactivator 1 alpha (219195_at), score: -0.53 PSMD5proteasome (prosome, macropain) 26S subunit, non-ATPase, 5 (203447_at), score: 0.64 PSTPIP2proline-serine-threonine phosphatase interacting protein 2 (219938_s_at), score: 0.69 PTPRDprotein tyrosine phosphatase, receptor type, D (214043_at), score: 0.65 RAD23BRAD23 homolog B (S. cerevisiae) (201222_s_at), score: 0.6 RASL11BRAS-like, family 11, member B (219142_at), score: 0.64 RECKreversion-inducing-cysteine-rich protein with kazal motifs (205407_at), score: 0.57 RGS4regulator of G-protein signaling 4 (204337_at), score: 0.57 RIPK4receptor-interacting serine-threonine kinase 4 (221215_s_at), score: -0.56 ROD1ROD1 regulator of differentiation 1 (S. pombe) (214697_s_at), score: 0.55 RPL30ribosomal protein L30 (200062_s_at), score: -0.52 RPRD1Aregulation of nuclear pre-mRNA domain containing 1A (218209_s_at), score: 0.6 RPS4P17ribosomal protein S4X pseudogene 17 (216342_x_at), score: -0.67 RPS4Xribosomal protein S4, X-linked (200933_x_at), score: -0.62 S100A4S100 calcium binding protein A4 (203186_s_at), score: 0.56 SCAND2SCAN domain containing 2 pseudogene (222177_s_at), score: -0.51 SCN3Asodium channel, voltage-gated, type III, alpha subunit (210432_s_at), score: 0.63 SCN9Asodium channel, voltage-gated, type IX, alpha subunit (206950_at), score: 0.62 SCRIBscribbled homolog (Drosophila) (212556_at), score: 0.66 SEMA4Csema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4C (46665_at), score: -0.5 SERPINB7serpin peptidase inhibitor, clade B (ovalbumin), member 7 (206421_s_at), score: 0.67 SERPINB9serpin peptidase inhibitor, clade B (ovalbumin), member 9 (209723_at), score: 0.83 SGK1serum/glucocorticoid regulated kinase 1 (201739_at), score: 0.62 SHROOM2shroom family member 2 (204967_at), score: 0.55 SIRT3sirtuin (silent mating type information regulation 2 homolog) 3 (S. cerevisiae) (221562_s_at), score: -0.53 SKP2S-phase kinase-associated protein 2 (p45) (203625_x_at), score: 0.56 SLC20A2solute carrier family 20 (phosphate transporter), member 2 (202744_at), score: 0.62 SLC24A1solute carrier family 24 (sodium/potassium/calcium exchanger), member 1 (206081_at), score: 0.61 SLC35F5solute carrier family 35, member F5 (220123_at), score: 0.64 SLC38A4solute carrier family 38, member 4 (220786_s_at), score: 0.91 SLC3A1solute carrier family 3 (cystine, dibasic and neutral amino acid transporters, activator of cystine, dibasic and neutral amino acid transport), member 1 (205799_s_at), score: 0.71 SLC48A1solute carrier family 48 (heme transporter), member 1 (218417_s_at), score: 0.58 SLFN12schlafen family member 12 (219885_at), score: 0.55 SMA5glucuronidase, beta pseudogene (215043_s_at), score: -0.59 SNRPD1small nuclear ribonucleoprotein D1 polypeptide 16kDa (202691_at), score: 0.55 SPCS3signal peptidase complex subunit 3 homolog (S. cerevisiae) (218817_at), score: 0.6 SPHARS-phase response (cyclin-related) (206272_at), score: 0.55 SQSTM1sequestosome 1 (201471_s_at), score: -0.52 ST6GAL1ST6 beta-galactosamide alpha-2,6-sialyltranferase 1 (201998_at), score: 0.61 STAM2signal transducing adaptor molecule (SH3 domain and ITAM motif) 2 (209649_at), score: 0.58 STAU2staufen, RNA binding protein, homolog 2 (Drosophila) (204226_at), score: 0.62 STOMstomatin (201060_x_at), score: 0.57 STX7syntaxin 7 (212631_at), score: 0.63 SULF1sulfatase 1 (212353_at), score: 0.6 SYNGR1synaptogyrin 1 (210613_s_at), score: 0.72 SYNMsynemin, intermediate filament protein (212730_at), score: 0.71 SYT11synaptotagmin XI (209198_s_at), score: 0.69 TBC1D2BTBC1 domain family, member 2B (212796_s_at), score: 0.59 TBXA2Rthromboxane A2 receptor (336_at), score: 0.91 TEStestis derived transcript (3 LIM domains) (202720_at), score: 0.62 TGFBItransforming growth factor, beta-induced, 68kDa (201506_at), score: -0.55 THAP1THAP domain containing, apoptosis associated protein 1 (219292_at), score: 0.62 THBS2thrombospondin 2 (203083_at), score: 0.62 TMEM111transmembrane protein 111 (217882_at), score: -0.62 TMEM135transmembrane protein 135 (222209_s_at), score: 0.56 TMEM168transmembrane protein 168 (218962_s_at), score: 0.65 TMEM2transmembrane protein 2 (218113_at), score: 0.72 TOXthymocyte selection-associated high mobility group box (204529_s_at), score: 0.65 TRHDEthyrotropin-releasing hormone degrading enzyme (219937_at), score: 0.63 TRIM32tripartite motif-containing 32 (203846_at), score: 0.55 TRNAG6transfer RNA glycine 6 (anticodon GCC) (217542_at), score: 0.63 TSKUtsukushin (218245_at), score: -0.56 TSPYL5TSPY-like 5 (213122_at), score: 0.61 TTTY15testis-specific transcript, Y-linked 15 (214983_at), score: 0.55 TUBB2Btubulin, beta 2B (214023_x_at), score: 0.57 TUBB6tubulin, beta 6 (209191_at), score: 0.55 UCP2uncoupling protein 2 (mitochondrial, proton carrier) (208998_at), score: 0.55 ULK2unc-51-like kinase 2 (C. elegans) (204062_s_at), score: 0.58 UTP18UTP18, small subunit (SSU) processome component, homolog (yeast) (203721_s_at), score: -0.5 VGLL3vestigial like 3 (Drosophila) (220327_at), score: 0.7 VPS13Avacuolar protein sorting 13 homolog A (S. cerevisiae) (214785_at), score: 0.57 WDR41WD repeat domain 41 (218055_s_at), score: 0.56 ZNF212zinc finger protein 212 (203985_at), score: -0.51 ZNF215zinc finger protein 215 (220214_at), score: 0.71 ZNF287zinc finger protein 287 (216710_x_at), score: -0.51 ZNF518Azinc finger protein 518A (204291_at), score: 0.65
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-GEOD-4219-raw-cel-1311956358.cel | 10 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956824.cel | 24 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956178.cel | 6 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |