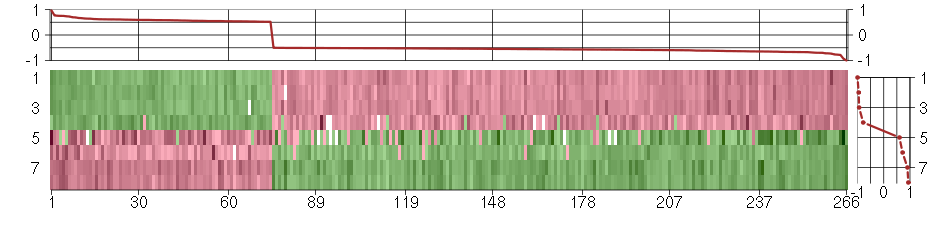

Under-expression is coded with green,
over-expression with red color.



ABLIM1actin binding LIM protein 1 (200965_s_at), score: -0.52 AEBP1AE binding protein 1 (201792_at), score: -0.65 AGPSalkylglycerone phosphate synthase (205401_at), score: 0.58 AHI1Abelson helper integration site 1 (221569_at), score: -0.57 AKAP12A kinase (PRKA) anchor protein 12 (210517_s_at), score: -0.59 AKAP2A kinase (PRKA) anchor protein 2 (202759_s_at), score: -0.51 AKR1B10aldo-keto reductase family 1, member B10 (aldose reductase) (206561_s_at), score: 0.76 ALDH1A1aldehyde dehydrogenase 1 family, member A1 (212224_at), score: -0.56 ALDH3A2aldehyde dehydrogenase 3 family, member A2 (202053_s_at), score: 0.56 ARF4ADP-ribosylation factor 4 (201096_s_at), score: -0.56 ARHGAP5Rho GTPase activating protein 5 (217936_at), score: -0.67 ARID4AAT rich interactive domain 4A (RBP1-like) (205062_x_at), score: -0.51 ARID5BAT rich interactive domain 5B (MRF1-like) (212614_at), score: -0.52 ARL15ADP-ribosylation factor-like 15 (219842_at), score: -0.55 ARMCX1armadillo repeat containing, X-linked 1 (218694_at), score: -0.67 ATF1activating transcription factor 1 (222103_at), score: -0.53 ATF7IP2activating transcription factor 7 interacting protein 2 (219870_at), score: -0.51 ATP1A1ATPase, Na+/K+ transporting, alpha 1 polypeptide (220948_s_at), score: 0.61 ATP2C1ATPase, Ca++ transporting, type 2C, member 1 (211137_s_at), score: -0.55 BAZ2Bbromodomain adjacent to zinc finger domain, 2B (203080_s_at), score: -0.6 BEX4brain expressed, X-linked 4 (215440_s_at), score: -0.51 BHLHB9basic helix-loop-helix domain containing, class B, 9 (213709_at), score: -0.51 BIRC3baculoviral IAP repeat-containing 3 (210538_s_at), score: 0.61 C14orf169chromosome 14 open reading frame 169 (219526_at), score: -0.64 C15orf29chromosome 15 open reading frame 29 (218791_s_at), score: -0.55 C16orf57chromosome 16 open reading frame 57 (218060_s_at), score: 0.54 C21orf59chromosome 21 open reading frame 59 (218123_at), score: 0.52 C5orf4chromosome 5 open reading frame 4 (48031_r_at), score: -0.64 C6orf120chromosome 6 open reading frame 120 (221786_at), score: -0.5 C6orf211chromosome 6 open reading frame 211 (218195_at), score: -0.52 C8orf4chromosome 8 open reading frame 4 (218541_s_at), score: -0.63 CAND1cullin-associated and neddylation-dissociated 1 (208839_s_at), score: -0.54 CAND2cullin-associated and neddylation-dissociated 2 (putative) (213547_at), score: -0.63 CCRL1chemokine (C-C motif) receptor-like 1 (220351_at), score: -0.51 CD24CD24 molecule (209771_x_at), score: 0.64 CD248CD248 molecule, endosialin (219025_at), score: -0.59 CDKN1Acyclin-dependent kinase inhibitor 1A (p21, Cip1) (202284_s_at), score: -0.63 CELSR1cadherin, EGF LAG seven-pass G-type receptor 1 (flamingo homolog, Drosophila) (41660_at), score: 0.62 CELSR3cadherin, EGF LAG seven-pass G-type receptor 3 (flamingo homolog, Drosophila) (40020_at), score: 0.77 CEP170centrosomal protein 170kDa (212746_s_at), score: -0.53 CHMP1Achromatin modifying protein 1A (201933_at), score: 0.6 CLEC11AC-type lectin domain family 11, member A (211709_s_at), score: -0.55 COL3A1collagen, type III, alpha 1 (215076_s_at), score: -0.68 CPEB3cytoplasmic polyadenylation element binding protein 3 (205773_at), score: -0.53 CSGALNACT2chondroitin sulfate N-acetylgalactosaminyltransferase 2 (222235_s_at), score: -0.58 CUGBP2CUG triplet repeat, RNA binding protein 2 (202158_s_at), score: -0.59 CYB5R2cytochrome b5 reductase 2 (220230_s_at), score: -0.57 DAPK1death-associated protein kinase 1 (203139_at), score: -0.52 DCNdecorin (211896_s_at), score: -0.59 DERL1Der1-like domain family, member 1 (219402_s_at), score: 0.59 DGKDdiacylglycerol kinase, delta 130kDa (208072_s_at), score: 0.72 DNAJC6DnaJ (Hsp40) homolog, subfamily C, member 6 (204720_s_at), score: -0.51 DNAJC7DnaJ (Hsp40) homolog, subfamily C, member 7 (202416_at), score: 0.6 DPY19L4dpy-19-like 4 (C. elegans) (213391_at), score: -0.57 DRAP1DR1-associated protein 1 (negative cofactor 2 alpha) (203258_at), score: 0.54 DRG1developmentally regulated GTP binding protein 1 (202810_at), score: 0.61 DSEdermatan sulfate epimerase (218854_at), score: -0.59 DZIP1DAZ interacting protein 1 (204557_s_at), score: -0.52 E2F1E2F transcription factor 1 (204947_at), score: 0.55 EIF2C2eukaryotic translation initiation factor 2C, 2 (213310_at), score: -0.55 EIF3Deukaryotic translation initiation factor 3, subunit D (200005_at), score: 0.58 EIF4A1eukaryotic translation initiation factor 4A, isoform 1 (211787_s_at), score: 0.65 EIF6eukaryotic translation initiation factor 6 (210213_s_at), score: 0.56 ELOVL4elongation of very long chain fatty acids (FEN1/Elo2, SUR4/Elo3, yeast)-like 4 (219532_at), score: -0.55 EMP1epithelial membrane protein 1 (201325_s_at), score: -0.61 ENOSF1enolase superfamily member 1 (204142_at), score: -0.51 ENPP2ectonucleotide pyrophosphatase/phosphodiesterase 2 (209392_at), score: -0.58 EPAS1endothelial PAS domain protein 1 (200878_at), score: -0.5 EPHX1epoxide hydrolase 1, microsomal (xenobiotic) (202017_at), score: 0.58 ERBB2IPerbb2 interacting protein (217941_s_at), score: -0.59 FAM174Bfamily with sequence similarity 174, member B (51158_at), score: -0.73 FAM65Bfamily with sequence similarity 65, member B (209829_at), score: -0.62 FCHSD2FCH and double SH3 domains 2 (203620_s_at), score: -0.55 FLJ10357hypothetical protein FLJ10357 (58780_s_at), score: -0.55 FLT1fms-related tyrosine kinase 1 (vascular endothelial growth factor/vascular permeability factor receptor) (222033_s_at), score: -0.62 FNDC3Afibronectin type III domain containing 3A (202304_at), score: -0.57 FOXG1forkhead box G1 (206018_at), score: 0.61 FOXL2forkhead box L2 (220102_at), score: 0.56 FOXO1forkhead box O1 (202724_s_at), score: -0.53 FZD4frizzled homolog 4 (Drosophila) (218665_at), score: -0.51 GAL3ST4galactose-3-O-sulfotransferase 4 (219815_at), score: -0.67 GALCgalactosylceramidase (204417_at), score: -0.7 GAS1growth arrest-specific 1 (204457_s_at), score: -0.57 GCC2GRIP and coiled-coil domain containing 2 (202832_at), score: -0.53 GIPC2GIPC PDZ domain containing family, member 2 (219970_at), score: -0.5 GNG12guanine nucleotide binding protein (G protein), gamma 12 (212294_at), score: -0.56 GOLPH3golgi phosphoprotein 3 (coat-protein) (217803_at), score: -0.58 GPR126G protein-coupled receptor 126 (213094_at), score: -0.53 GRAMD3GRAM domain containing 3 (218706_s_at), score: -0.55 GREM1gremlin 1, cysteine knot superfamily, homolog (Xenopus laevis) (218469_at), score: -1 HOXA10homeobox A10 (213150_at), score: -0.57 HSD17B12hydroxysteroid (17-beta) dehydrogenase 12 (217869_at), score: -0.67 HSD17B7hydroxysteroid (17-beta) dehydrogenase 7 (220081_x_at), score: 0.54 HYPKHuntingtin interacting protein K (218680_x_at), score: 0.66 IGSF3immunoglobulin superfamily, member 3 (202421_at), score: 0.75 IL10RBinterleukin 10 receptor, beta (209575_at), score: 0.53 IL1R1interleukin 1 receptor, type I (202948_at), score: -0.55 IL23Ainterleukin 23, alpha subunit p19 (211796_s_at), score: -0.62 ILF2interleukin enhancer binding factor 2, 45kDa (200052_s_at), score: 0.57 IPWimprinted in Prader-Willi syndrome (non-protein coding) (221974_at), score: -0.67 IRAK3interleukin-1 receptor-associated kinase 3 (213817_at), score: -0.51 ISL1ISL LIM homeobox 1 (206104_at), score: -0.64 KARSlysyl-tRNA synthetase (200079_s_at), score: 0.53 KYNUkynureninase (L-kynurenine hydrolase) (217388_s_at), score: 0.96 LAS1LLAS1-like (S. cerevisiae) (208117_s_at), score: 0.56 LEF1lymphoid enhancer-binding factor 1 (221558_s_at), score: 0.69 LGMNlegumain (201212_at), score: -0.55 LIPAlipase A, lysosomal acid, cholesterol esterase (201847_at), score: -0.52 LMO7LIM domain 7 (202674_s_at), score: -0.52 LOC728344similar to hCG1978918 (216532_x_at), score: 0.57 LOC728855hypothetical LOC728855 (222001_x_at), score: 0.63 LPPR4plasticity related gene 1 (213496_at), score: -0.65 LRIG1leucine-rich repeats and immunoglobulin-like domains 1 (211596_s_at), score: -0.57 LRRC2leucine rich repeat containing 2 (219949_at), score: -0.56 LSG1large subunit GTPase 1 homolog (S. cerevisiae) (221536_s_at), score: 0.54 LTBP2latent transforming growth factor beta binding protein 2 (204682_at), score: -0.6 LUMlumican (201744_s_at), score: -0.93 LXNlatexin (218729_at), score: -0.59 MAPRE3microtubule-associated protein, RP/EB family, member 3 (203842_s_at), score: 0.58 MED23mediator complex subunit 23 (218846_at), score: -0.61 MEIS1Meis homeobox 1 (204069_at), score: -0.51 MEIS3P1Meis homeobox 3 pseudogene 1 (214077_x_at), score: -0.66 MLF1myeloid leukemia factor 1 (204784_s_at), score: 0.55 MLLT3myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 3 (204918_s_at), score: -0.63 MORC3MORC family CW-type zinc finger 3 (213000_at), score: -0.54 MOXD1monooxygenase, DBH-like 1 (209708_at), score: -0.6 MPPED2metallophosphoesterase domain containing 2 (205413_at), score: 0.73 MRPL28mitochondrial ribosomal protein L28 (204599_s_at), score: 0.52 MXRA5matrix-remodelling associated 5 (209596_at), score: -0.54 MYO1Bmyosin IB (212364_at), score: -0.77 MYO6myosin VI (203216_s_at), score: -0.52 NASPnuclear autoantigenic sperm protein (histone-binding) (201970_s_at), score: 0.58 NBEAneurobeachin (221207_s_at), score: -0.53 NDNnecdin homolog (mouse) (209550_at), score: -0.54 NEK7NIMA (never in mitosis gene a)-related kinase 7 (212530_at), score: -0.61 NESnestin (218678_at), score: -0.56 NLGN1neuroligin 1 (205893_at), score: -0.53 NPAS2neuronal PAS domain protein 2 (39549_at), score: -0.59 NPTNneuroplastin (202228_s_at), score: -0.53 NPTX1neuronal pentraxin I (204684_at), score: 0.59 NRP1neuropilin 1 (212298_at), score: -0.54 NT5E5'-nucleotidase, ecto (CD73) (203939_at), score: -0.51 NUDT4nudix (nucleoside diphosphate linked moiety X)-type motif 4 (212183_at), score: -0.52 OR2A9Polfactory receptor, family 2, subfamily A, member 9 pseudogene (222290_at), score: -0.54 P2RY5purinergic receptor P2Y, G-protein coupled, 5 (218589_at), score: -0.58 PBX1pre-B-cell leukemia homeobox 1 (212148_at), score: -0.54 PBXIP1pre-B-cell leukemia homeobox interacting protein 1 (214176_s_at), score: -0.57 PCLOpiccolo (presynaptic cytomatrix protein) (213558_at), score: 0.75 PCNXpecanex homolog (Drosophila) (213173_at), score: -0.54 PCTK2PCTAIRE protein kinase 2 (221918_at), score: -0.72 PDGFRAplatelet-derived growth factor receptor, alpha polypeptide (203131_at), score: -0.76 PDLIM4PDZ and LIM domain 4 (214175_x_at), score: -0.52 PEX5peroxisomal biogenesis factor 5 (203244_at), score: 0.54 PFDN2prefoldin subunit 2 (218336_at), score: 0.69 PGAP1post-GPI attachment to proteins 1 (213469_at), score: -0.59 PID1phosphotyrosine interaction domain containing 1 (219093_at), score: -0.54 PJA2praja ring finger 2 (201133_s_at), score: -0.65 PKIGprotein kinase (cAMP-dependent, catalytic) inhibitor gamma (202732_at), score: -0.56 PLAG1pleiomorphic adenoma gene 1 (205372_at), score: -0.58 PLAGL1pleiomorphic adenoma gene-like 1 (209318_x_at), score: -0.57 PLCB1phospholipase C, beta 1 (phosphoinositide-specific) (213222_at), score: -0.64 PLCG1phospholipase C, gamma 1 (202789_at), score: 0.58 PLK2polo-like kinase 2 (Drosophila) (201939_at), score: 0.55 POSTNperiostin, osteoblast specific factor (210809_s_at), score: -0.52 PPIHpeptidylprolyl isomerase H (cyclophilin H) (204228_at), score: 0.6 PPP2R2Bprotein phosphatase 2 (formerly 2A), regulatory subunit B, beta isoform (213849_s_at), score: 0.53 PRDM2PR domain containing 2, with ZNF domain (203057_s_at), score: -0.51 PRR16proline rich 16 (220014_at), score: -0.65 PRRG1proline rich Gla (G-carboxyglutamic acid) 1 (205618_at), score: -0.54 PRRX1paired related homeobox 1 (205991_s_at), score: -0.51 PSMB6proteasome (prosome, macropain) subunit, beta type, 6 (208827_at), score: 0.62 PSMB8proteasome (prosome, macropain) subunit, beta type, 8 (large multifunctional peptidase 7) (209040_s_at), score: 0.52 PSMD13proteasome (prosome, macropain) 26S subunit, non-ATPase, 13 (201232_s_at), score: 0.52 PSMD4proteasome (prosome, macropain) 26S subunit, non-ATPase, 4 (211609_x_at), score: 0.6 PSMD5proteasome (prosome, macropain) 26S subunit, non-ATPase, 5 (203447_at), score: -0.51 PSTPIP2proline-serine-threonine phosphatase interacting protein 2 (219938_s_at), score: -0.72 PTPRDprotein tyrosine phosphatase, receptor type, D (214043_at), score: -0.67 PUM2pumilio homolog 2 (Drosophila) (216221_s_at), score: -0.51 RAB33BRAB33B, member RAS oncogene family (221014_s_at), score: -0.55 RECKreversion-inducing-cysteine-rich protein with kazal motifs (205407_at), score: -0.67 RGP1RGP1 retrograde golgi transport homolog (S. cerevisiae) (203169_at), score: 0.55 RGS4regulator of G-protein signaling 4 (204337_at), score: -0.64 RIPK4receptor-interacting serine-threonine kinase 4 (221215_s_at), score: 0.64 RND3Rho family GTPase 3 (212724_at), score: -0.57 ROCK1Rho-associated, coiled-coil containing protein kinase 1 (213044_at), score: -0.55 RP2retinitis pigmentosa 2 (X-linked recessive) (205191_at), score: -0.6 RPRD1Aregulation of nuclear pre-mRNA domain containing 1A (218209_s_at), score: -0.51 RPS3ribosomal protein S3 (208692_at), score: 0.58 RPS6KA1ribosomal protein S6 kinase, 90kDa, polypeptide 1 (203379_at), score: 0.55 RUNX1runt-related transcription factor 1 (209360_s_at), score: -0.57 S100A4S100 calcium binding protein A4 (203186_s_at), score: -0.51 SALL2sal-like 2 (Drosophila) (213283_s_at), score: -0.57 SAMD4Asterile alpha motif domain containing 4A (212845_at), score: -0.52 SAV1salvador homolog 1 (Drosophila) (218276_s_at), score: -0.55 SCRIBscribbled homolog (Drosophila) (212556_at), score: -0.53 SEMA3Fsema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3F (35666_at), score: -0.57 SENP6SUMO1/sentrin specific peptidase 6 (202318_s_at), score: -0.51 SERINC1serine incorporator 1 (208671_at), score: -0.51 SERPINB9serpin peptidase inhibitor, clade B (ovalbumin), member 9 (209723_at), score: -0.64 SGK1serum/glucocorticoid regulated kinase 1 (201739_at), score: -0.59 SH3BGRSH3 domain binding glutamic acid-rich protein (204979_s_at), score: 0.55 SIM1single-minded homolog 1 (Drosophila) (206876_at), score: -0.52 SLC25A5solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 5 (200657_at), score: 0.55 SLC30A1solute carrier family 30 (zinc transporter), member 1 (212907_at), score: -0.65 SLC38A4solute carrier family 38, member 4 (220786_s_at), score: -0.57 SLC3A1solute carrier family 3 (cystine, dibasic and neutral amino acid transporters, activator of cystine, dibasic and neutral amino acid transport), member 1 (205799_s_at), score: -0.51 SMA4glucuronidase, beta pseudogene (215599_at), score: 0.59 SMA5glucuronidase, beta pseudogene (215043_s_at), score: 0.59 SMAD4SMAD family member 4 (202527_s_at), score: -0.56 SMAP1small ArfGAP 1 (218137_s_at), score: -0.53 SMARCE1SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily e, member 1 (211989_at), score: 0.59 SNF8SNF8, ESCRT-II complex subunit, homolog (S. cerevisiae) (218391_at), score: 0.61 SOCS5suppressor of cytokine signaling 5 (209647_s_at), score: -0.62 SPANXCSPANX family, member C (220217_x_at), score: 0.67 SPHARS-phase response (cyclin-related) (206272_at), score: -0.59 SSPNsarcospan (Kras oncogene-associated gene) (204963_at), score: -0.53 SSRP1structure specific recognition protein 1 (200956_s_at), score: 0.59 STAM2signal transducing adaptor molecule (SH3 domain and ITAM motif) 2 (209649_at), score: -0.62 STIM1stromal interaction molecule 1 (202764_at), score: 0.56 STK38Lserine/threonine kinase 38 like (212572_at), score: -0.55 STOMstomatin (201060_x_at), score: -0.54 STRN3striatin, calmodulin binding protein 3 (204496_at), score: -0.51 STX12syntaxin 12 (212112_s_at), score: -0.54 SULF1sulfatase 1 (212353_at), score: -0.65 SYNGR1synaptogyrin 1 (210613_s_at), score: -0.67 TBC1D9TBC1 domain family, member 9 (with GRAM domain) (212956_at), score: -0.58 TBXA2Rthromboxane A2 receptor (336_at), score: -0.65 TCF12transcription factor 12 (208986_at), score: -0.57 TCF7L2transcription factor 7-like 2 (T-cell specific, HMG-box) (212761_at), score: -0.63 TEStestis derived transcript (3 LIM domains) (202720_at), score: -0.78 TGFB2transforming growth factor, beta 2 (220407_s_at), score: 0.61 TGFBItransforming growth factor, beta-induced, 68kDa (201506_at), score: 0.53 THAP1THAP domain containing, apoptosis associated protein 1 (219292_at), score: -0.51 THBS2thrombospondin 2 (203083_at), score: -0.7 TMEM111transmembrane protein 111 (217882_at), score: 0.54 TMEM168transmembrane protein 168 (218962_s_at), score: -0.52 TMEM186transmembrane protein 186 (204676_at), score: 0.52 TMEM2transmembrane protein 2 (218113_at), score: -0.54 TMEM30Atransmembrane protein 30A (217743_s_at), score: -0.66 TMEM50Atransmembrane protein 50A (217766_s_at), score: -0.51 TNS1tensin 1 (221748_s_at), score: -0.7 TOB1transducer of ERBB2, 1 (202704_at), score: -0.56 TOR3Atorsin family 3, member A (218459_at), score: 0.57 TRPC3transient receptor potential cation channel, subfamily C, member 3 (210814_at), score: 0.62 TRPS1trichorhinophalangeal syndrome I (218502_s_at), score: -0.58 TSPAN13tetraspanin 13 (217979_at), score: -0.52 TSPAN6tetraspanin 6 (209108_at), score: -0.53 TSPYL4TSPY-like 4 (212928_at), score: -0.51 TSPYL5TSPY-like 5 (213122_at), score: -0.69 TTC33tetratricopeptide repeat domain 33 (219421_at), score: -0.51 TTLL12tubulin tyrosine ligase-like family, member 12 (216251_s_at), score: 0.53 U2AF1U2 small nuclear RNA auxiliary factor 1 (202858_at), score: 0.58 UBE2Bubiquitin-conjugating enzyme E2B (RAD6 homolog) (211763_s_at), score: -0.5 ULK2unc-51-like kinase 2 (C. elegans) (204062_s_at), score: -0.61 UTP18UTP18, small subunit (SSU) processome component, homolog (yeast) (203721_s_at), score: 0.59 UTYubiquitously transcribed tetratricopeptide repeat gene, Y-linked (211149_at), score: -0.56 VGLL3vestigial like 3 (Drosophila) (220327_at), score: -0.6 YIPF4Yip1 domain family, member 4 (209551_at), score: -0.52 YPEL5yippee-like 5 (Drosophila) (217783_s_at), score: -0.65 ZBTB10zinc finger and BTB domain containing 10 (219312_s_at), score: -0.52 ZBTB6zinc finger and BTB domain containing 6 (206098_at), score: -0.53 ZCCHC6zinc finger, CCHC domain containing 6 (220933_s_at), score: -0.57 ZDHHC17zinc finger, DHHC-type containing 17 (212982_at), score: -0.61 ZHX2zinc fingers and homeoboxes 2 (203556_at), score: -0.57 ZNF215zinc finger protein 215 (220214_at), score: -0.57 ZNF518Azinc finger protein 518A (204291_at), score: -0.6
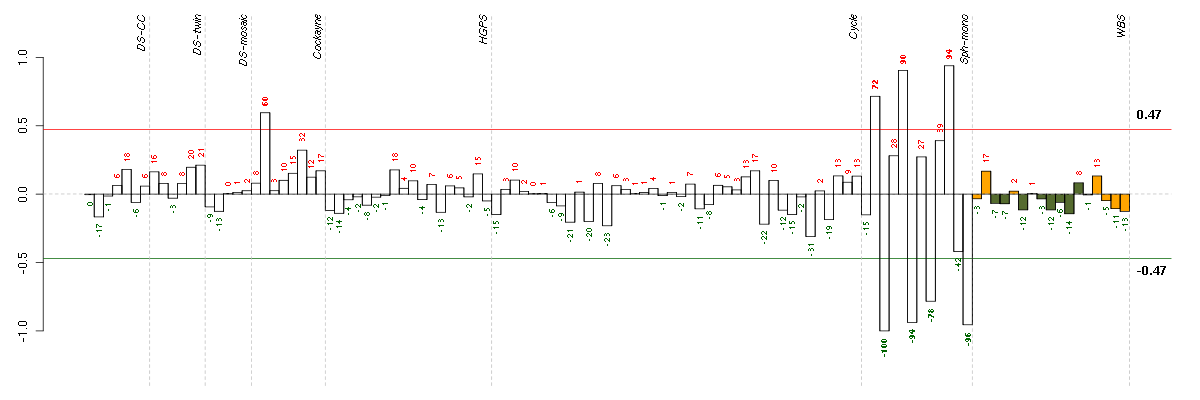
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-GEOD-4219-raw-cel-1311956178.cel | 6 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956824.cel | 24 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956358.cel | 10 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956418.cel | 13 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-3407-raw-cel-1437949579.cel | 2 | 4 | Cockayne | hgu133a | none | CSB |
| E-GEOD-4219-raw-cel-1311956138.cel | 4 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956321.cel | 9 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956614.cel | 18 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |