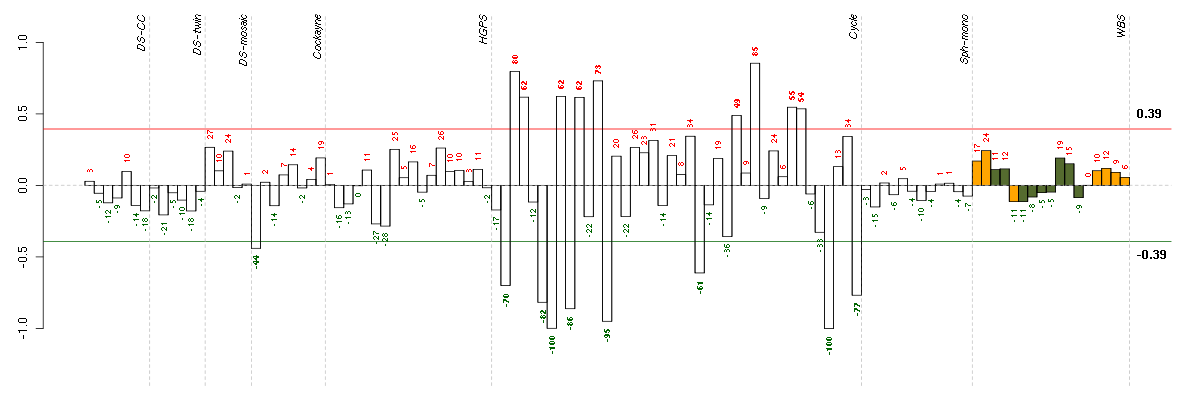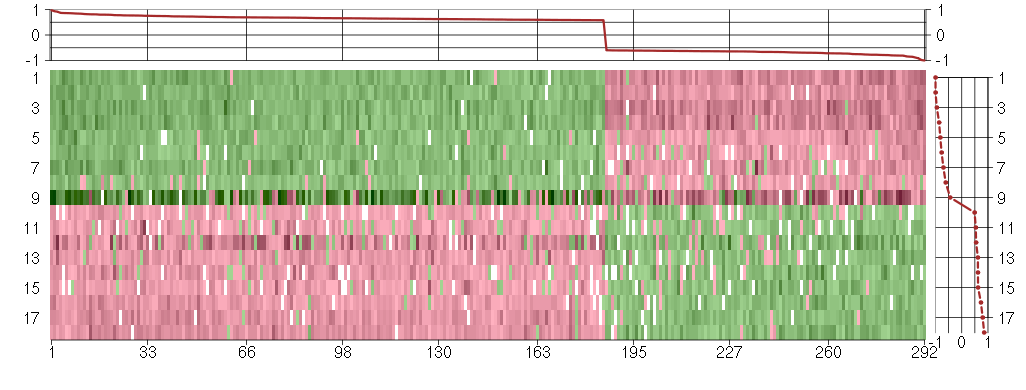

Under-expression is coded with green,
over-expression with red color.

biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
positive regulation of kinase activity
Any process that activates or increases the frequency, rate or extent of kinase activity, the catalysis of the transfer of a phosphate group, usually from ATP, to a substrate molecule.
positive regulation of catalytic activity
Any process that activates or increases the activity of an enzyme.
regulation of MAP kinase activity
Any process that modulates the frequency, rate or extent of MAP kinase activity.
positive regulation of MAP kinase activity
Any process that activates or increases the frequency, rate or extent of MAP kinase activity.
regulation of JUN kinase activity
Any process that modulates the frequency, rate or extent of JUN kinase activity.
positive regulation of JUN kinase activity
Any process that activates or increases the frequency, rate or extent of JUN kinase activity.
regulation of kinase activity
Any process that modulates the frequency, rate or extent of kinase activity, the catalysis of the transfer of a phosphate group, usually from ATP, to a substrate molecule.
regulation of protein kinase activity
Any process that modulates the frequency, rate or extent of protein kinase activity.
positive regulation of protein kinase activity
Any process that activates or increases the frequency, rate or extent of protein kinase activity.
regulation of catalytic activity
Any process that modulates the activity of an enzyme.
regulation of transferase activity
Any process that modulates the frequency, rate or extent of transferase activity, the catalysis of the transfer of a group, e.g. a methyl group, glycosyl group, acyl group, phosphorus-containing, or other groups, from one compound (generally regarded as the donor) to another compound (generally regarded as the acceptor). Transferase is the systematic name for any enzyme of EC class 2.
positive regulation of transferase activity
Any process that activates or increases the frequency, rate or extent of transferase activity, the catalysis of the transfer of a group, e.g. a methyl group, glycosyl group, acyl group, phosphorus-containing, or other groups, from a donor compound to an acceptor.
biological regulation
Any process that modulates the frequency, rate or extent of any biological process, quality or function.
regulation of molecular function
Any process that modulates the frequency, rate or extent of molecular functions. Molecular functions are elemental biological activities occurring at the molecular level, such as catalysis or binding.
all
This term is the most general term possible
positive regulation of transferase activity
Any process that activates or increases the frequency, rate or extent of transferase activity, the catalysis of the transfer of a group, e.g. a methyl group, glycosyl group, acyl group, phosphorus-containing, or other groups, from a donor compound to an acceptor.
positive regulation of kinase activity
Any process that activates or increases the frequency, rate or extent of kinase activity, the catalysis of the transfer of a phosphate group, usually from ATP, to a substrate molecule.
positive regulation of protein kinase activity
Any process that activates or increases the frequency, rate or extent of protein kinase activity.
positive regulation of MAP kinase activity
Any process that activates or increases the frequency, rate or extent of MAP kinase activity.
positive regulation of JUN kinase activity
Any process that activates or increases the frequency, rate or extent of JUN kinase activity.


AAASachalasia, adrenocortical insufficiency, alacrimia (Allgrove, triple-A) (218075_at), score: 0.64 ABCA7ATP-binding cassette, sub-family A (ABC1), member 7 (219577_s_at), score: -0.66 ABCA8ATP-binding cassette, sub-family A (ABC1), member 8 (204719_at), score: 0.62 ABCD1ATP-binding cassette, sub-family D (ALD), member 1 (205142_x_at), score: 0.73 ABLIM1actin binding LIM protein 1 (200965_s_at), score: 0.65 ACSL4acyl-CoA synthetase long-chain family member 4 (202422_s_at), score: -0.72 ACTC1actin, alpha, cardiac muscle 1 (205132_at), score: -0.64 ACTN3actinin, alpha 3 (206891_at), score: -0.63 ACTR3BARP3 actin-related protein 3 homolog B (yeast) (218868_at), score: -0.72 ADAMTS12ADAM metallopeptidase with thrombospondin type 1 motif, 12 (221421_s_at), score: -0.62 ADARadenosine deaminase, RNA-specific (201786_s_at), score: 0.72 ADCY3adenylate cyclase 3 (209321_s_at), score: 0.61 ADORA2Badenosine A2b receptor (205891_at), score: 0.66 AGPAT11-acylglycerol-3-phosphate O-acyltransferase 1 (lysophosphatidic acid acyltransferase, alpha) (215535_s_at), score: 0.73 AGRNagrin (217419_x_at), score: 0.61 AIM1absent in melanoma 1 (212543_at), score: 0.65 AKAP2A kinase (PRKA) anchor protein 2 (202759_s_at), score: 0.64 ALDH1L1aldehyde dehydrogenase 1 family, member L1 (215798_at), score: -0.81 ANXA10annexin A10 (210143_at), score: -0.61 ARSAarylsulfatase A (204443_at), score: 0.6 ATN1atrophin 1 (40489_at), score: 0.61 ATP2C1ATPase, Ca++ transporting, type 2C, member 1 (211137_s_at), score: -0.73 B9D1B9 protein domain 1 (210534_s_at), score: 0.63 BAP1BRCA1 associated protein-1 (ubiquitin carboxy-terminal hydrolase) (201419_at), score: 0.7 BAT2HLA-B associated transcript 2 (212081_x_at), score: 0.8 BAT2LHLA-B associated transcript 2-like (212068_s_at), score: 0.65 BCL2L1BCL2-like 1 (215037_s_at), score: 0.63 BDKRB2bradykinin receptor B2 (205870_at), score: 0.58 BIN1bridging integrator 1 (210201_x_at), score: 0.59 BMP6bone morphogenetic protein 6 (206176_at), score: -0.64 BTBD2BTB (POZ) domain containing 2 (207722_s_at), score: 0.75 BTG2BTG family, member 2 (201236_s_at), score: 0.68 C10orf97chromosome 10 open reading frame 97 (218297_at), score: -0.64 C16orf45chromosome 16 open reading frame 45 (212736_at), score: 0.59 C4orf43chromosome 4 open reading frame 43 (218513_at), score: -0.72 C5orf44chromosome 5 open reading frame 44 (218674_at), score: -0.61 CALB2calbindin 2 (205428_s_at), score: -0.85 CALCOCO1calcium binding and coiled-coil domain 1 (209002_s_at), score: 0.79 CAPN5calpain 5 (205166_at), score: 0.59 CASC3cancer susceptibility candidate 3 (207842_s_at), score: 0.62 CCDC101coiled-coil domain containing 101 (48117_at), score: -0.61 CD248CD248 molecule, endosialin (219025_at), score: 0.6 CDC25Bcell division cycle 25 homolog B (S. pombe) (201853_s_at), score: 0.62 CDC42BPBCDC42 binding protein kinase beta (DMPK-like) (217849_s_at), score: 0.81 CDC42EP1CDC42 effector protein (Rho GTPase binding) 1 (204693_at), score: 0.62 CDC42EP4CDC42 effector protein (Rho GTPase binding) 4 (214721_x_at), score: 0.82 CDH6cadherin 6, type 2, K-cadherin (fetal kidney) (205532_s_at), score: -0.62 CHMP5chromatin modifying protein 5 (218085_at), score: -0.61 CHUKconserved helix-loop-helix ubiquitous kinase (209666_s_at), score: -0.64 CICcapicua homolog (Drosophila) (212784_at), score: 0.8 CITcitron (rho-interacting, serine/threonine kinase 21) (212801_at), score: 0.59 CIZ1CDKN1A interacting zinc finger protein 1 (205516_x_at), score: 0.7 CLIP3CAP-GLY domain containing linker protein 3 (212358_at), score: 0.67 CNIH3cornichon homolog 3 (Drosophila) (214841_at), score: 0.62 CNOT3CCR4-NOT transcription complex, subunit 3 (203239_s_at), score: 0.77 COL18A1collagen, type XVIII, alpha 1 (209081_s_at), score: 0.66 CRTC3CREB regulated transcription coactivator 3 (218648_at), score: 0.86 CScitrate synthase (208660_at), score: 0.72 CUL7cullin 7 (36084_at), score: 0.59 DAXXdeath-domain associated protein (201763_s_at), score: 0.58 DDB2damage-specific DNA binding protein 2, 48kDa (203409_at), score: 0.63 DENND2ADENN/MADD domain containing 2A (53991_at), score: 0.69 DENND5BDENN/MADD domain containing 5B (215058_at), score: -0.62 DERL2Der1-like domain family, member 2 (218333_at), score: -0.61 DKFZP564O0823DKFZP564O0823 protein (204687_at), score: -0.71 DMPKdystrophia myotonica-protein kinase (37996_s_at), score: 0.82 DNAJB9DnaJ (Hsp40) homolog, subfamily B, member 9 (202843_at), score: -0.78 DNAJC6DnaJ (Hsp40) homolog, subfamily C, member 6 (204720_s_at), score: -0.69 DOPEY1dopey family member 1 (40612_at), score: -0.66 DZIP3DAZ interacting protein 3, zinc finger (213186_at), score: 0.6 ECM1extracellular matrix protein 1 (209365_s_at), score: 0.62 EDEM1ER degradation enhancer, mannosidase alpha-like 1 (203279_at), score: -0.64 EHMT2euchromatic histone-lysine N-methyltransferase 2 (202326_at), score: 0.61 ENTPD1ectonucleoside triphosphate diphosphohydrolase 1 (209473_at), score: -0.73 EPHA4EPH receptor A4 (206114_at), score: -0.79 ERBB2IPerbb2 interacting protein (217941_s_at), score: -0.64 FAM102Afamily with sequence similarity 102, member A (212400_at), score: 0.61 FAM18Bfamily with sequence similarity 18, member B (218446_s_at), score: -0.72 FAM62Afamily with sequence similarity 62 (C2 domain containing), member A (208858_s_at), score: 0.6 FBXL14F-box and leucine-rich repeat protein 14 (213145_at), score: 0.83 FBXO17F-box protein 17 (220233_at), score: 0.7 FCF1FCF1 small subunit (SSU) processome component homolog (S. cerevisiae) (219927_at), score: -0.65 FGGYFGGY carbohydrate kinase domain containing (219718_at), score: 0.65 FICDFIC domain containing (219910_at), score: -0.64 FLJ12529pre-mRNA cleavage factor I, 59 kDa subunit (217866_at), score: 0.62 FNBP1formin binding protein 1 (212288_at), score: 0.6 FOLR3folate receptor 3 (gamma) (206371_at), score: -0.68 FOXC2forkhead box C2 (MFH-1, mesenchyme forkhead 1) (214520_at), score: 0.74 FOXF1forkhead box F1 (205935_at), score: 0.68 FOXK2forkhead box K2 (203064_s_at), score: 0.86 FRG1FSHD region gene 1 (204145_at), score: -0.61 GABARAPL3GABA(A) receptors associated protein like 3 (pseudogene) (211458_s_at), score: -0.62 GFOD1glucose-fructose oxidoreductase domain containing 1 (219821_s_at), score: -0.75 GKglycerol kinase (207387_s_at), score: -0.62 GK3Pglycerol kinase 3 pseudogene (215966_x_at), score: -0.77 GMCL1germ cell-less homolog 1 (Drosophila) (218458_at), score: -0.61 GMPRguanosine monophosphate reductase (204187_at), score: 0.77 GOLT1Bgolgi transport 1 homolog B (S. cerevisiae) (218193_s_at), score: -0.69 GPR56G protein-coupled receptor 56 (212070_at), score: 0.6 GTF2A1Lgeneral transcription factor IIA, 1-like (213413_at), score: 0.67 H1F0H1 histone family, member 0 (208886_at), score: 0.58 H1FXH1 histone family, member X (204805_s_at), score: 0.74 HAB1B1 for mucin (215778_x_at), score: -0.79 HAMPhepcidin antimicrobial peptide (220491_at), score: -0.7 HDGFhepatoma-derived growth factor (high-mobility group protein 1-like) (200896_x_at), score: 0.62 HECTD3HECT domain containing 3 (218632_at), score: 0.59 HGFhepatocyte growth factor (hepapoietin A; scatter factor) (209960_at), score: 0.68 HIST1H4Ghistone cluster 1, H4g (208551_at), score: -0.61 HLA-DQB1major histocompatibility complex, class II, DQ beta 1 (211654_x_at), score: -0.65 HMGA1high mobility group AT-hook 1 (210457_x_at), score: 0.72 HNRNPUL1heterogeneous nuclear ribonucleoprotein U-like 1 (209675_s_at), score: 0.64 HSD17B6hydroxysteroid (17-beta) dehydrogenase 6 homolog (mouse) (37512_at), score: -0.88 HSPA12Aheat shock 70kDa protein 12A (214434_at), score: 0.7 HSPA4Lheat shock 70kDa protein 4-like (205543_at), score: 0.59 IDUAiduronidase, alpha-L- (205059_s_at), score: 0.69 INE1inactivation escape 1 (non-protein coding) (207252_at), score: -0.67 IRF1interferon regulatory factor 1 (202531_at), score: 0.76 ITGA1integrin, alpha 1 (214660_at), score: -0.64 JUNDjun D proto-oncogene (203751_x_at), score: 0.73 JUPjunction plakoglobin (201015_s_at), score: 0.93 KAL1Kallmann syndrome 1 sequence (205206_at), score: -0.98 KIAA1305KIAA1305 (220911_s_at), score: 0.91 KLHL23kelch-like 23 (Drosophila) (213610_s_at), score: -0.8 KPNA6karyopherin alpha 6 (importin alpha 7) (212101_at), score: 0.61 LARP4La ribonucleoprotein domain family, member 4 (214155_s_at), score: -0.6 LEPREL2leprecan-like 2 (204854_at), score: 0.62 LIN7Blin-7 homolog B (C. elegans) (219760_at), score: -0.61 LLGL2lethal giant larvae homolog 2 (Drosophila) (203713_s_at), score: -0.64 LOC100132540similar to LOC339047 protein (214870_x_at), score: 0.58 LOC100133105hypothetical protein LOC100133105 (214237_x_at), score: -0.66 LOC339047hypothetical protein LOC339047 (221501_x_at), score: 0.66 LOC391132similar to hCG2041276 (216177_at), score: -0.65 LOC440434hypothetical protein FLJ11822 (215090_x_at), score: 0.72 LPGAT1lysophosphatidylglycerol acyltransferase 1 (202651_at), score: -0.67 LRDDleucine-rich repeats and death domain containing (221640_s_at), score: 0.69 LRP2low density lipoprotein-related protein 2 (205710_at), score: 0.68 LRP8low density lipoprotein receptor-related protein 8, apolipoprotein e receptor (205282_at), score: -0.85 MAN1C1mannosidase, alpha, class 1C, member 1 (218918_at), score: 0.69 MAP1Smicrotubule-associated protein 1S (218522_s_at), score: 0.66 MAP2K5mitogen-activated protein kinase kinase 5 (211370_s_at), score: 0.63 MAP3K6mitogen-activated protein kinase kinase kinase 6 (219278_at), score: 0.63 MAP4microtubule-associated protein 4 (200836_s_at), score: 0.6 MAST4microtubule associated serine/threonine kinase family member 4 (40016_g_at), score: 0.66 MDM2Mdm2 p53 binding protein homolog (mouse) (205386_s_at), score: 0.68 MFAP3Lmicrofibrillar-associated protein 3-like (205442_at), score: -0.66 MGAT1mannosyl (alpha-1,3-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase (201126_s_at), score: 0.64 MINK1misshapen-like kinase 1 (zebrafish) (214246_x_at), score: 0.62 MLL4myeloid/lymphoid or mixed-lineage leukemia 4 (203419_at), score: 0.59 MMP10matrix metallopeptidase 10 (stromelysin 2) (205680_at), score: -0.78 MMP16matrix metallopeptidase 16 (membrane-inserted) (207012_at), score: -0.61 MPP1membrane protein, palmitoylated 1, 55kDa (202974_at), score: 0.69 MUC1mucin 1, cell surface associated (207847_s_at), score: 0.67 MUL1mitochondrial E3 ubiquitin ligase 1 (218246_at), score: 0.66 MYL4myosin, light chain 4, alkali; atrial, embryonic (210088_x_at), score: -0.81 NARG1LNMDA receptor regulated 1-like (219378_at), score: -0.63 NCALDneurocalcin delta (211685_s_at), score: -0.64 NCKAP1NCK-associated protein 1 (217465_at), score: -0.73 NDPNorrie disease (pseudoglioma) (206022_at), score: 0.78 NEFLneurofilament, light polypeptide (221805_at), score: -0.66 NEFMneurofilament, medium polypeptide (205113_at), score: -1 NF1neurofibromin 1 (211094_s_at), score: 0.68 NFATC4nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 4 (205897_at), score: 0.6 NFKBIAnuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha (201502_s_at), score: 0.76 NINJ1ninjurin 1 (203045_at), score: 0.59 NOL10nucleolar protein 10 (218591_s_at), score: -0.63 NOVA2neuro-oncological ventral antigen 2 (206477_s_at), score: -0.63 NPIPnuclear pore complex interacting protein (204538_x_at), score: 0.71 NUPL2nucleoporin like 2 (204003_s_at), score: -0.64 NXPH3neurexophilin 3 (221991_at), score: -0.66 OAS22'-5'-oligoadenylate synthetase 2, 69/71kDa (204972_at), score: 0.68 OAS32'-5'-oligoadenylate synthetase 3, 100kDa (218400_at), score: 0.71 OBFC2Boligonucleotide/oligosaccharide-binding fold containing 2B (218903_s_at), score: 0.6 OLFML2Bolfactomedin-like 2B (213125_at), score: 0.81 OLR1oxidized low density lipoprotein (lectin-like) receptor 1 (210004_at), score: -0.69 OPCMLopioid binding protein/cell adhesion molecule-like (214111_at), score: -0.64 OSTM1osteopetrosis associated transmembrane protein 1 (218196_at), score: -0.77 P2RY5purinergic receptor P2Y, G-protein coupled, 5 (218589_at), score: 0.69 PAK1p21 protein (Cdc42/Rac)-activated kinase 1 (209615_s_at), score: 0.59 PARNpoly(A)-specific ribonuclease (deadenylation nuclease) (203905_at), score: 0.62 PARVBparvin, beta (204629_at), score: 0.66 PCDH9protocadherin 9 (219737_s_at), score: -0.91 PCDHG@protocadherin gamma cluster (215836_s_at), score: 0.64 PCDHGA1protocadherin gamma subfamily A, 1 (209079_x_at), score: 0.64 PCDHGA11protocadherin gamma subfamily A, 11 (211876_x_at), score: 0.62 PCDHGA3protocadherin gamma subfamily A, 3 (216352_x_at), score: 0.64 PCYT2phosphate cytidylyltransferase 2, ethanolamine (209577_at), score: 0.69 PDE5Aphosphodiesterase 5A, cGMP-specific (206757_at), score: 0.8 PDE7Bphosphodiesterase 7B (220343_at), score: -0.62 PHF7PHD finger protein 7 (215622_x_at), score: -0.71 PI4K2Aphosphatidylinositol 4-kinase type 2 alpha (209345_s_at), score: 0.59 PIGLphosphatidylinositol glycan anchor biosynthesis, class L (213889_at), score: 0.61 PIP5K1Cphosphatidylinositol-4-phosphate 5-kinase, type I, gamma (212518_at), score: 0.77 PITX1paired-like homeodomain 1 (208502_s_at), score: 0.69 PKN1protein kinase N1 (202161_at), score: 0.7 PLAUplasminogen activator, urokinase (211668_s_at), score: 0.73 PLCD1phospholipase C, delta 1 (205125_at), score: 0.59 POLR2Apolymerase (RNA) II (DNA directed) polypeptide A, 220kDa (202725_at), score: 0.68 POMZP3POM (POM121 homolog, rat) and ZP3 fusion (204148_s_at), score: 0.68 PPARGperoxisome proliferator-activated receptor gamma (208510_s_at), score: 0.71 PRMT3protein arginine methyltransferase 3 (213320_at), score: -0.7 PTPROprotein tyrosine phosphatase, receptor type, O (211600_at), score: 0.75 PXNpaxillin (211823_s_at), score: 0.67 RAB11BRAB11B, member RAS oncogene family (34478_at), score: 0.6 RAB1ARAB1A, member RAS oncogene family (213440_at), score: -0.8 RBM15BRNA binding motif protein 15B (202689_at), score: 0.69 REREarginine-glutamic acid dipeptide (RE) repeats (200940_s_at), score: 0.85 RFX5regulatory factor X, 5 (influences HLA class II expression) (202963_at), score: 0.65 RNF11ring finger protein 11 (208924_at), score: -0.63 RNF220ring finger protein 220 (219988_s_at), score: 0.77 ROR2receptor tyrosine kinase-like orphan receptor 2 (205578_at), score: 0.67 RP11-345P4.4similar to solute carrier family 35, member E2 (217122_s_at), score: 0.62 RP5-1000E10.4suppressor of IKK epsilon (221705_s_at), score: -0.72 RPL18AP6ribosomal protein L18a pseudogene 6 (216383_at), score: -0.84 RPLP2P1ribosomal protein, large P2, pseudogene 1 (216490_x_at), score: -0.67 RPRD2regulation of nuclear pre-mRNA domain containing 2 (212553_at), score: 0.66 RPS17P5ribosomal protein S17 pseudogene 5 (216348_at), score: -0.76 RRP15ribosomal RNA processing 15 homolog (S. cerevisiae) (219037_at), score: -0.62 SAP30LSAP30-like (219129_s_at), score: 0.6 SAV1salvador homolog 1 (Drosophila) (218276_s_at), score: 0.84 SBNO2strawberry notch homolog 2 (Drosophila) (204166_at), score: 0.72 SCAMP4secretory carrier membrane protein 4 (213244_at), score: 0.97 SDAD1SDA1 domain containing 1 (218607_s_at), score: 0.63 SDF2L1stromal cell-derived factor 2-like 1 (218681_s_at), score: -0.64 SDPRserum deprivation response (phosphatidylserine binding protein) (218711_s_at), score: 0.72 SEMA3Fsema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3F (35666_at), score: -0.63 SF1splicing factor 1 (208313_s_at), score: 0.79 SH3GLB2SH3-domain GRB2-like endophilin B2 (218813_s_at), score: 0.66 SIGIRRsingle immunoglobulin and toll-interleukin 1 receptor (TIR) domain (52940_at), score: 0.74 SIGLEC6sialic acid binding Ig-like lectin 6 (210796_x_at), score: -0.7 SIRT6sirtuin (silent mating type information regulation 2 homolog) 6 (S. cerevisiae) (219613_s_at), score: 0.62 SLC25A13solute carrier family 25, member 13 (citrin) (203775_at), score: -0.75 SLC2A6solute carrier family 2 (facilitated glucose transporter), member 6 (220091_at), score: 0.58 SLC4A7solute carrier family 4, sodium bicarbonate cotransporter, member 7 (209884_s_at), score: -0.77 SLC5A5solute carrier family 5 (sodium iodide symporter), member 5 (211123_at), score: -0.75 SLC6A11solute carrier family 6 (neurotransmitter transporter, GABA), member 11 (207048_at), score: -0.68 SLC8A1solute carrier family 8 (sodium/calcium exchanger), member 1 (207053_at), score: -0.6 SMAD3SMAD family member 3 (218284_at), score: 0.81 SMG6Smg-6 homolog, nonsense mediated mRNA decay factor (C. elegans) (214940_s_at), score: 0.66 SMG7Smg-7 homolog, nonsense mediated mRNA decay factor (C. elegans) (201794_s_at), score: 0.58 SMN1survival of motor neuron 1, telomeric (203852_s_at), score: -0.64 SNAP25synaptosomal-associated protein, 25kDa (202508_s_at), score: 0.75 SOLHsmall optic lobes homolog (Drosophila) (204275_at), score: 0.65 SORBS1sorbin and SH3 domain containing 1 (218087_s_at), score: -0.61 SPATA20spermatogenesis associated 20 (218164_at), score: 0.61 SPG7spastic paraplegia 7 (pure and complicated autosomal recessive) (202104_s_at), score: 0.64 ST5suppression of tumorigenicity 5 (202440_s_at), score: 0.66 STIM1stromal interaction molecule 1 (202764_at), score: 0.73 SULT1B1sulfotransferase family, cytosolic, 1B, member 1 (207601_at), score: -0.7 SYNE1spectrin repeat containing, nuclear envelope 1 (209447_at), score: -0.67 SYNPOsynaptopodin (202796_at), score: 0.58 TAF13TAF13 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 18kDa (205966_at), score: -0.7 TAPBPTAP binding protein (tapasin) (208829_at), score: 0.69 TBC1D2BTBC1 domain family, member 2B (212796_s_at), score: 0.68 TENC1tensin like C1 domain containing phosphatase (tensin 2) (212494_at), score: 0.75 TERF2telomeric repeat binding factor 2 (203611_at), score: 0.65 TGDSTDP-glucose 4,6-dehydratase (208249_s_at), score: -0.62 THSD7Athrombospondin, type I, domain containing 7A (214920_at), score: 0.62 TIAF1TGFB1-induced anti-apoptotic factor 1 (202039_at), score: 0.74 TICAM2toll-like receptor adaptor molecule 2 (214658_at), score: -0.77 TLE3transducin-like enhancer of split 3 (E(sp1) homolog, Drosophila) (206472_s_at), score: 0.63 TLE4transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila) (204872_at), score: -0.63 TLR3toll-like receptor 3 (206271_at), score: 0.63 TNIP1TNFAIP3 interacting protein 1 (207196_s_at), score: 0.64 TP53tumor protein p53 (201746_at), score: 0.86 TRIM21tripartite motif-containing 21 (204804_at), score: 0.78 TRMT2ATRM2 tRNA methyltransferase 2 homolog A (S. cerevisiae) (91617_at), score: -0.69 TRPC4transient receptor potential cation channel, subfamily C, member 4 (220817_at), score: -0.63 TSC2tuberous sclerosis 2 (215735_s_at), score: 0.69 TSKUtsukushin (218245_at), score: 0.84 TTC13tetratricopeptide repeat domain 13 (219481_at), score: -0.62 TUBA4Btubulin, alpha 4b (pseudogene) (207490_at), score: -0.81 TXLNAtaxilin alpha (212300_at), score: 0.63 UBAC1UBA domain containing 1 (202151_s_at), score: 0.6 UBTD1ubiquitin domain containing 1 (219172_at), score: 0.68 ULBP2UL16 binding protein 2 (221291_at), score: -0.75 ULK1unc-51-like kinase 1 (C. elegans) (209333_at), score: 0.66 USF2upstream transcription factor 2, c-fos interacting (202152_x_at), score: 0.64 VEGFBvascular endothelial growth factor B (203683_s_at), score: 0.78 VPS37Bvacuolar protein sorting 37 homolog B (S. cerevisiae) (221704_s_at), score: 0.72 VWA5Avon Willebrand factor A domain containing 5A (210102_at), score: 0.58 WBP2WW domain binding protein 2 (209117_at), score: 0.69 WDR4WD repeat domain 4 (221632_s_at), score: -0.68 WDR42AWD repeat domain 42A (202249_s_at), score: 0.76 WDR6WD repeat domain 6 (217734_s_at), score: 0.87 YAF2YY1 associated factor 2 (206238_s_at), score: -0.63 YES1v-yes-1 Yamaguchi sarcoma viral oncogene homolog 1 (202932_at), score: 0.75 YPEL1yippee-like 1 (Drosophila) (206063_x_at), score: -0.64 ZCCHC10zinc finger, CCHC domain containing 10 (221193_s_at), score: -0.6 ZDHHC4zinc finger, DHHC-type containing 4 (220261_s_at), score: 0.63 ZDHHC7zinc finger, DHHC-type containing 7 (218606_at), score: 0.7 ZNF580zinc finger protein 580 (220748_s_at), score: 0.65 ZNF706zinc finger protein 706 (218059_at), score: 0.6
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-TABM-263-raw-cel-1515486371.cel | 37 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485771.cel | 7 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485891.cel | 13 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485811.cel | 9 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485751.cel | 6 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486431.cel | 40 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485671.cel | 2 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486091.cel | 23 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-GEOD-3407-raw-cel-1437949557.cel | 1 | 4 | Cockayne | hgu133a | CS | eGFP |
| E-TABM-263-raw-cel-1515486171.cel | 27 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486311.cel | 34 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486291.cel | 33 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485831.cel | 10 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485711.cel | 4 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485791.cel | 8 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485871.cel | 12 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485691.cel | 3 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486211.cel | 29 | 6 | Cycle | hgu133a2 | none | Cycle 1 |