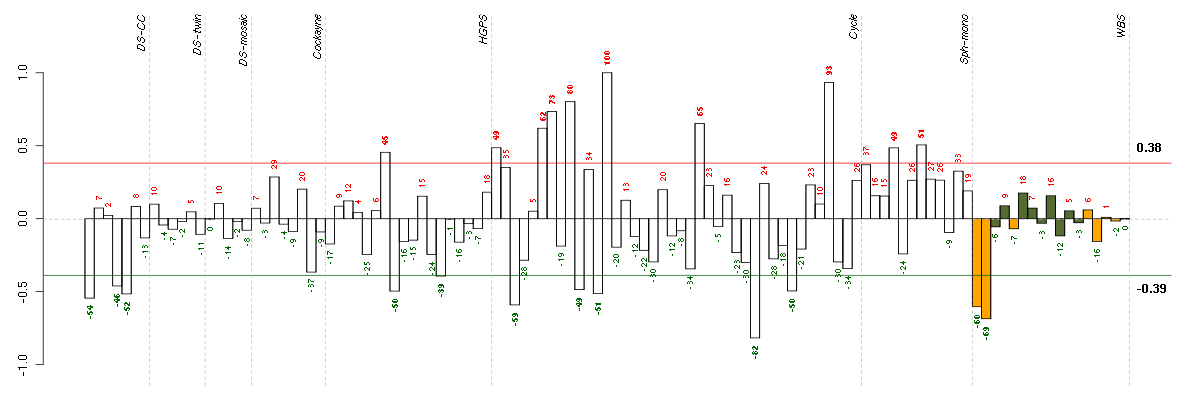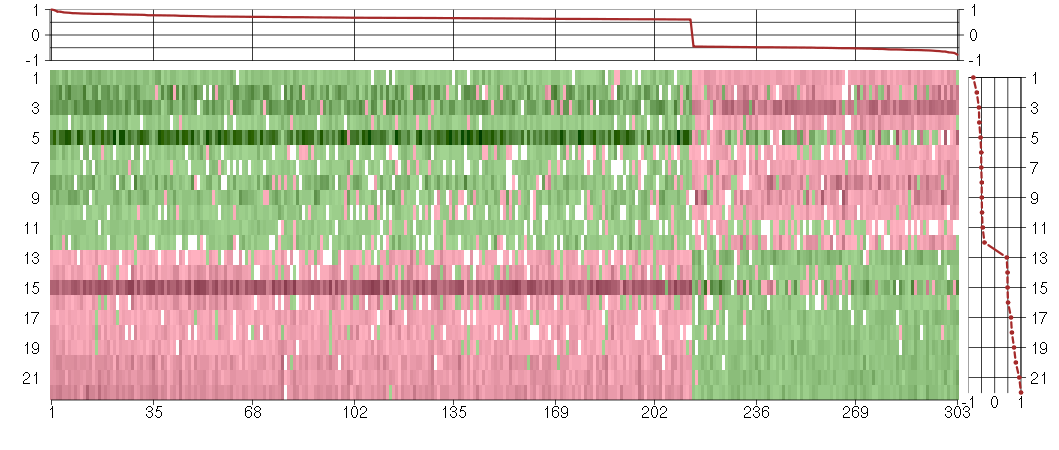

Under-expression is coded with green,
over-expression with red color.



AAASachalasia, adrenocortical insufficiency, alacrimia (Allgrove, triple-A) (218075_at), score: -0.59 ABCD1ATP-binding cassette, sub-family D (ALD), member 1 (205142_x_at), score: -0.53 ABCE1ATP-binding cassette, sub-family E (OABP), member 1 (201873_s_at), score: 0.83 ACAP2ArfGAP with coiled-coil, ankyrin repeat and PH domains 2 (212476_at), score: 0.71 ACSL1acyl-CoA synthetase long-chain family member 1 (201963_at), score: 0.74 ACSL4acyl-CoA synthetase long-chain family member 4 (202422_s_at), score: 0.84 ACTR3BARP3 actin-related protein 3 homolog B (yeast) (218868_at), score: 0.67 ADCK2aarF domain containing kinase 2 (221893_s_at), score: -0.53 AGPAT11-acylglycerol-3-phosphate O-acyltransferase 1 (lysophosphatidic acid acyltransferase, alpha) (215535_s_at), score: -0.58 AHCTF1AT hook containing transcription factor 1 (214766_s_at), score: 0.62 AHI1Abelson helper integration site 1 (221569_at), score: 0.62 AP2A2adaptor-related protein complex 2, alpha 2 subunit (212159_x_at), score: -0.48 APBB1amyloid beta (A4) precursor protein-binding, family B, member 1 (Fe65) (202652_at), score: -0.57 ARFGEF2ADP-ribosylation factor guanine nucleotide-exchange factor 2 (brefeldin A-inhibited) (218098_at), score: 0.66 ARSAarylsulfatase A (204443_at), score: -0.61 ASAP3ArfGAP with SH3 domain, ankyrin repeat and PH domain 3 (219103_at), score: -0.46 ATF2activating transcription factor 2 (212984_at), score: 0.68 ATN1atrophin 1 (40489_at), score: -0.55 ATP2C1ATPase, Ca++ transporting, type 2C, member 1 (211137_s_at), score: 0.7 ATP8B1ATPase, class I, type 8B, member 1 (214594_x_at), score: 0.72 ATXN2Lataxin 2-like (201806_s_at), score: -0.46 BAT3HLA-B associated transcript 3 (213318_s_at), score: -0.49 BBS10Bardet-Biedl syndrome 10 (219487_at), score: 0.67 BCAP29B-cell receptor-associated protein 29 (205084_at), score: 0.62 BMI1BMI1 polycomb ring finger oncogene (202265_at), score: 0.67 BNIP2BCL2/adenovirus E1B 19kDa interacting protein 2 (209308_s_at), score: 0.72 BRD4bromodomain containing 4 (202102_s_at), score: -0.52 BTBD2BTB (POZ) domain containing 2 (207722_s_at), score: -0.71 C10orf97chromosome 10 open reading frame 97 (218297_at), score: 0.63 C12orf29chromosome 12 open reading frame 29 (213701_at), score: 0.68 C14orf138chromosome 14 open reading frame 138 (218940_at), score: 0.65 C14orf147chromosome 14 open reading frame 147 (212460_at), score: 0.62 C14orf94chromosome 14 open reading frame 94 (218383_at), score: -0.53 C1orf109chromosome 1 open reading frame 109 (218712_at), score: 0.65 C1orf9chromosome 1 open reading frame 9 (203429_s_at), score: 0.67 C20orf149chromosome 20 open reading frame 149 (218010_x_at), score: -0.53 C3orf18chromosome 3 open reading frame 18 (219114_at), score: -0.51 C4orf43chromosome 4 open reading frame 43 (218513_at), score: 0.83 C5orf22chromosome 5 open reading frame 22 (203738_at), score: 0.68 C5orf28chromosome 5 open reading frame 28 (219029_at), score: 0.66 C5orf44chromosome 5 open reading frame 44 (218674_at), score: 0.82 C6orf211chromosome 6 open reading frame 211 (218195_at), score: 0.67 C7orf64chromosome 7 open reading frame 64 (221596_s_at), score: 0.73 C9orf82chromosome 9 open reading frame 82 (219276_x_at), score: 0.66 CA9carbonic anhydrase IX (205199_at), score: -0.49 CABIN1calcineurin binding protein 1 (37652_at), score: -0.47 CALCOCO1calcium binding and coiled-coil domain 1 (209002_s_at), score: -0.65 CAPZA2capping protein (actin filament) muscle Z-line, alpha 2 (201237_at), score: 0.71 CASP7caspase 7, apoptosis-related cysteine peptidase (207181_s_at), score: 0.63 CD248CD248 molecule, endosialin (219025_at), score: -0.57 CD2APCD2-associated protein (203593_at), score: 0.68 CDC42BPBCDC42 binding protein kinase beta (DMPK-like) (217849_s_at), score: -0.55 CDC42EP1CDC42 effector protein (Rho GTPase binding) 1 (204693_at), score: -0.51 CDC73cell division cycle 73, Paf1/RNA polymerase II complex component, homolog (S. cerevisiae) (218578_at), score: 0.73 CDH6cadherin 6, type 2, K-cadherin (fetal kidney) (205532_s_at), score: 0.61 CEBPZCCAAT/enhancer binding protein (C/EBP), zeta (203341_at), score: 0.7 CHMP5chromatin modifying protein 5 (218085_at), score: 0.81 CHORDC1cysteine and histidine-rich domain (CHORD)-containing 1 (218566_s_at), score: 0.84 CHUKconserved helix-loop-helix ubiquitous kinase (209666_s_at), score: 0.86 CICcapicua homolog (Drosophila) (212784_at), score: -0.68 CIZ1CDKN1A interacting zinc finger protein 1 (205516_x_at), score: -0.49 CLN5ceroid-lipofuscinosis, neuronal 5 (204084_s_at), score: 0.65 CMPK1cytidine monophosphate (UMP-CMP) kinase 1, cytosolic (217870_s_at), score: 0.83 CNOT3CCR4-NOT transcription complex, subunit 3 (203239_s_at), score: -0.5 CRCPCGRP receptor component (203898_at), score: 0.7 CSGALNACT2chondroitin sulfate N-acetylgalactosaminyltransferase 2 (222235_s_at), score: 0.72 CUL7cullin 7 (36084_at), score: -0.47 DBN1drebrin 1 (217025_s_at), score: -0.49 DCP2DCP2 decapping enzyme homolog (S. cerevisiae) (212919_at), score: 0.7 DCUN1D4DCN1, defective in cullin neddylation 1, domain containing 4 (S. cerevisiae) (212855_at), score: 0.63 DDX52DEAD (Asp-Glu-Ala-Asp) box polypeptide 52 (212834_at), score: 0.62 DERL2Der1-like domain family, member 2 (218333_at), score: 0.7 DHX29DEAH (Asp-Glu-Ala-His) box polypeptide 29 (212648_at), score: 0.66 DNAJB14DnaJ (Hsp40) homolog, subfamily B, member 14 (219237_s_at), score: 0.63 DNAJB9DnaJ (Hsp40) homolog, subfamily B, member 9 (202843_at), score: 0.77 DNAJC24DnaJ (Hsp40) homolog, subfamily C, member 24 (213853_at), score: 0.72 DNM1dynamin 1 (215116_s_at), score: -0.52 DOCK9dedicator of cytokinesis 9 (212538_at), score: 0.67 DOPEY1dopey family member 1 (40612_at), score: 0.67 DUSP4dual specificity phosphatase 4 (204014_at), score: 0.64 EAF2ELL associated factor 2 (219551_at), score: 0.62 ECM1extracellular matrix protein 1 (209365_s_at), score: -0.5 EDEM1ER degradation enhancer, mannosidase alpha-like 1 (203279_at), score: 0.61 EIF2AK3eukaryotic translation initiation factor 2-alpha kinase 3 (218696_at), score: 0.67 EIF2S1eukaryotic translation initiation factor 2, subunit 1 alpha, 35kDa (201143_s_at), score: 0.66 EIF3Meukaryotic translation initiation factor 3, subunit M (202231_at), score: 0.68 EIF4Eeukaryotic translation initiation factor 4E (201436_at), score: 0.85 EIF5Beukaryotic translation initiation factor 5B (214314_s_at), score: 0.65 EPS15epidermal growth factor receptor pathway substrate 15 (217887_s_at), score: 0.66 ERBB2IPerbb2 interacting protein (217941_s_at), score: 0.71 EVI5ecotropic viral integration site 5 (209717_at), score: 0.64 FAM18Bfamily with sequence similarity 18, member B (218446_s_at), score: 0.97 FAM29Afamily with sequence similarity 29, member A (218602_s_at), score: 0.63 FAM3Cfamily with sequence similarity 3, member C (201889_at), score: 0.68 FAM49Bfamily with sequence similarity 49, member B (217916_s_at), score: 0.65 FBLN2fibulin 2 (203886_s_at), score: -0.49 FKBP8FK506 binding protein 8, 38kDa (208255_s_at), score: -0.51 FLOT1flotillin 1 (210142_x_at), score: -0.49 FMR1fragile X mental retardation 1 (215245_x_at), score: 0.68 FPGTfucose-1-phosphate guanylyltransferase (205140_at), score: 0.63 FRG1FSHD region gene 1 (204145_at), score: 0.77 FUBP3far upstream element (FUSE) binding protein 3 (212824_at), score: 0.78 G3BP2GTPase activating protein (SH3 domain) binding protein 2 (208841_s_at), score: 0.81 GFOD1glucose-fructose oxidoreductase domain containing 1 (219821_s_at), score: 0.68 GMCL1germ cell-less homolog 1 (Drosophila) (218458_at), score: 0.75 GNG11guanine nucleotide binding protein (G protein), gamma 11 (204115_at), score: 0.62 GNPTABN-acetylglucosamine-1-phosphate transferase, alpha and beta subunits (212959_s_at), score: 0.62 GOLT1Bgolgi transport 1 homolog B (S. cerevisiae) (218193_s_at), score: 0.65 GRINAglutamate receptor, ionotropic, N-methyl D-aspartate-associated protein 1 (glutamate binding) (212090_at), score: -0.48 GRSF1G-rich RNA sequence binding factor 1 (201501_s_at), score: 0.71 GTF2F2general transcription factor IIF, polypeptide 2, 30kDa (209595_at), score: 0.66 HBS1LHBS1-like (S. cerevisiae) (209314_s_at), score: 0.62 HDAC2histone deacetylase 2 (201833_at), score: 0.66 HDAC6histone deacetylase 6 (206846_s_at), score: -0.48 HDAC7histone deacetylase 7 (217937_s_at), score: -0.46 HISPPD1histidine acid phosphatase domain containing 1 (203253_s_at), score: 0.87 HMGA1high mobility group AT-hook 1 (210457_x_at), score: -0.46 HNRNPH2heterogeneous nuclear ribonucleoprotein H2 (H') (201132_at), score: 0.8 HOXA4homeobox A4 (206289_at), score: -0.47 HSD17B6hydroxysteroid (17-beta) dehydrogenase 6 homolog (mouse) (37512_at), score: 0.7 HSPA9heat shock 70kDa protein 9 (mortalin) (200690_at), score: 0.61 HSPB6heat shock protein, alpha-crystallin-related, B6 (214767_s_at), score: -0.59 HUS1HUS1 checkpoint homolog (S. pombe) (217618_x_at), score: 0.61 IBTKinhibitor of Bruton agammaglobulinemia tyrosine kinase (210970_s_at), score: 0.69 IFRD1interferon-related developmental regulator 1 (202146_at), score: 0.71 IMPACTImpact homolog (mouse) (218637_at), score: 0.68 IRS1insulin receptor substrate 1 (204686_at), score: 0.64 ITSN2intersectin 2 (209898_x_at), score: 0.61 JARID1Cjumonji, AT rich interactive domain 1C (202383_at), score: -0.45 JMJD1Cjumonji domain containing 1C (221763_at), score: 0.71 JUNDjun D proto-oncogene (203751_x_at), score: -0.65 JUPjunction plakoglobin (201015_s_at), score: -0.47 KAL1Kallmann syndrome 1 sequence (205206_at), score: 0.7 KIAA0494KIAA0494 (201776_s_at), score: 0.66 KIAA0562KIAA0562 (204075_s_at), score: 0.77 KIAA1305KIAA1305 (220911_s_at), score: -0.52 KLHL2kelch-like 2, Mayven (Drosophila) (219157_at), score: 0.61 LARP4La ribonucleoprotein domain family, member 4 (214155_s_at), score: 0.92 LDLRlow density lipoprotein receptor (202068_s_at), score: -0.46 LOC100132540similar to LOC339047 protein (214870_x_at), score: -0.59 LOC286434hypothetical protein LOC286434 (222196_at), score: -0.46 LOC339047hypothetical protein LOC339047 (221501_x_at), score: -0.48 LOC391132similar to hCG2041276 (216177_at), score: 0.64 LOC440354PI-3-kinase-related kinase SMG-1 pseudogene (210396_s_at), score: 0.73 LPGAT1lysophosphatidylglycerol acyltransferase 1 (202651_at), score: 0.84 LRDDleucine-rich repeats and death domain containing (221640_s_at), score: -0.48 LRP8low density lipoprotein receptor-related protein 8, apolipoprotein e receptor (205282_at), score: 0.71 LRRC40leucine rich repeat containing 40 (218577_at), score: 0.81 LTBP3latent transforming growth factor beta binding protein 3 (219922_s_at), score: -0.48 MAP1Smicrotubule-associated protein 1S (218522_s_at), score: -0.59 MAP2K2mitogen-activated protein kinase kinase 2 (213490_s_at), score: -0.48 MAP2K5mitogen-activated protein kinase kinase 5 (211370_s_at), score: -0.5 MAP3K6mitogen-activated protein kinase kinase kinase 6 (219278_at), score: -0.5 MAP7D1MAP7 domain containing 1 (217943_s_at), score: -0.52 MAPK1IP1Lmitogen-activated protein kinase 1 interacting protein 1-like (212499_s_at), score: 0.7 MATR3matrin 3 (200626_s_at), score: 0.76 MED23mediator complex subunit 23 (218846_at), score: 0.66 MED4mediator complex subunit 4 (217843_s_at), score: 0.67 MEX3Cmex-3 homolog C (C. elegans) (218247_s_at), score: 0.65 MFAP3microfibrillar-associated protein 3 (213123_at), score: 0.65 MFFmitochondrial fission factor (219137_s_at), score: 0.66 MINK1misshapen-like kinase 1 (zebrafish) (214246_x_at), score: -0.56 MIS12MIS12, MIND kinetochore complex component, homolog (yeast) (221559_s_at), score: 0.66 MORF4L1mortality factor 4 like 1 (217982_s_at), score: 0.62 MOSPD1motile sperm domain containing 1 (218853_s_at), score: 0.65 MPHOSPH10M-phase phosphoprotein 10 (U3 small nucleolar ribonucleoprotein) (212885_at), score: 0.62 MRS2MRS2 magnesium homeostasis factor homolog (S. cerevisiae) (218538_s_at), score: 0.61 MUC1mucin 1, cell surface associated (207847_s_at), score: -0.69 NARG1LNMDA receptor regulated 1-like (219378_at), score: 0.73 NAT13N-acetyltransferase 13 (GCN5-related) (217745_s_at), score: 0.71 NCKAP1NCK-associated protein 1 (217465_at), score: 0.67 NDUFA5NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 5, 13kDa (201304_at), score: 0.83 NFATC4nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 4 (205897_at), score: -0.58 NFKBIAnuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha (201502_s_at), score: -0.47 NPATnuclear protein, ataxia-telangiectasia locus (209798_at), score: 0.68 NPIPnuclear pore complex interacting protein (204538_x_at), score: -0.6 NRN1neuritin 1 (218625_at), score: -0.46 NUP160nucleoporin 160kDa (212709_at), score: 0.79 NUPL2nucleoporin like 2 (204003_s_at), score: 0.62 NXT2nuclear transport factor 2-like export factor 2 (209628_at), score: 0.62 OLR1oxidized low density lipoprotein (lectin-like) receptor 1 (210004_at), score: 0.62 OPA1optic atrophy 1 (autosomal dominant) (212213_x_at), score: 0.76 OSTM1osteopetrosis associated transmembrane protein 1 (218196_at), score: 0.89 PAQR3progestin and adipoQ receptor family member III (213372_at), score: 0.8 PCDHGA1protocadherin gamma subfamily A, 1 (209079_x_at), score: -0.48 PCNPPEST proteolytic signal containing nuclear protein (217816_s_at), score: 0.63 PCTK2PCTAIRE protein kinase 2 (221918_at), score: 0.63 PDCLphosducin-like (204449_at), score: 0.63 PDK2pyruvate dehydrogenase kinase, isozyme 2 (202590_s_at), score: -0.46 PEX6peroxisomal biogenesis factor 6 (204545_at), score: -0.63 PFKLphosphofructokinase, liver (201102_s_at), score: -0.47 PIP5K1Cphosphatidylinositol-4-phosphate 5-kinase, type I, gamma (212518_at), score: -0.51 PKD1polycystic kidney disease 1 (autosomal dominant) (202328_s_at), score: -0.5 PLCD1phospholipase C, delta 1 (205125_at), score: -0.62 PLD3phospholipase D family, member 3 (201050_at), score: -0.54 PLSCR3phospholipid scramblase 3 (218828_at), score: -0.49 PNPLA2patatin-like phospholipase domain containing 2 (212705_x_at), score: -0.77 POLR2Apolymerase (RNA) II (DNA directed) polypeptide A, 220kDa (202725_at), score: -0.53 PPATphosphoribosyl pyrophosphate amidotransferase (209433_s_at), score: 0.67 PPFIA1protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 1 (202066_at), score: 0.65 PPP1CBprotein phosphatase 1, catalytic subunit, beta isoform (201407_s_at), score: 0.67 PPP3R1protein phosphatase 3 (formerly 2B), regulatory subunit B, alpha isoform (204507_s_at), score: 0.69 PRKCDprotein kinase C, delta (202545_at), score: -0.47 PRMT3protein arginine methyltransferase 3 (213320_at), score: 0.83 PRPS2phosphoribosyl pyrophosphate synthetase 2 (203401_at), score: 0.61 PTK2PTK2 protein tyrosine kinase 2 (208820_at), score: 0.63 PTOV1prostate tumor overexpressed 1 (212032_s_at), score: -0.57 PVRL3poliovirus receptor-related 3 (213325_at), score: 0.65 RAB1ARAB1A, member RAS oncogene family (213440_at), score: 0.82 RAB7ARAB7A, member RAS oncogene family (211960_s_at), score: 0.76 RABGGTBRab geranylgeranyltransferase, beta subunit (213704_at), score: 0.67 RANBP6RAN binding protein 6 (213019_at), score: 0.85 RB1CC1RB1-inducible coiled-coil 1 (202034_x_at), score: 0.67 RBM15BRNA binding motif protein 15B (202689_at), score: -0.63 RECQLRecQ protein-like (DNA helicase Q1-like) (205091_x_at), score: 0.67 RIF1RAP1 interacting factor homolog (yeast) (214700_x_at), score: 0.64 RINT1RAD50 interactor 1 (218598_at), score: 0.63 RNF11ring finger protein 11 (208924_at), score: 0.77 RNF138ring finger protein 138 (218738_s_at), score: 0.64 RP2retinitis pigmentosa 2 (X-linked recessive) (205191_at), score: 0.71 RP5-1000E10.4suppressor of IKK epsilon (221705_s_at), score: 0.68 RPS10P11ribosomal protein S10 pseudogene 11 (216505_x_at), score: 0.65 RPS17P5ribosomal protein S17 pseudogene 5 (216348_at), score: 0.61 RPS25ribosomal protein S25 (200091_s_at), score: 0.69 RRP15ribosomal RNA processing 15 homolog (S. cerevisiae) (219037_at), score: 0.8 SACM1LSAC1 suppressor of actin mutations 1-like (yeast) (202797_at), score: 0.75 SBNO2strawberry notch homolog 2 (Drosophila) (204166_at), score: -0.54 SCAMP4secretory carrier membrane protein 4 (213244_at), score: -0.53 SEC24BSEC24 family, member B (S. cerevisiae) (202798_at), score: 0.65 SEC62SEC62 homolog (S. cerevisiae) (208942_s_at), score: 0.67 SEP1515 kDa selenoprotein (200902_at), score: 0.67 SF3A2splicing factor 3a, subunit 2, 66kDa (37462_i_at), score: -0.48 SLC25A13solute carrier family 25, member 13 (citrin) (203775_at), score: 0.72 SLC25A24solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 24 (204342_at), score: 0.61 SLC25A32solute carrier family 25, member 32 (221020_s_at), score: 0.69 SLC2A4RGSLC2A4 regulator (218494_s_at), score: -0.49 SLC30A9solute carrier family 30 (zinc transporter), member 9 (202614_at), score: 0.71 SLC33A1solute carrier family 33 (acetyl-CoA transporter), member 1 (203165_s_at), score: 0.66 SLC4A7solute carrier family 4, sodium bicarbonate cotransporter, member 7 (209884_s_at), score: 0.81 SLMO2slowmo homolog 2 (Drosophila) (217851_s_at), score: 0.92 SLTMSAFB-like, transcription modulator (217828_at), score: 0.62 SMCHD1structural maintenance of chromosomes flexible hinge domain containing 1 (212569_at), score: 0.7 SMN1survival of motor neuron 1, telomeric (203852_s_at), score: 0.69 SNRNP27small nuclear ribonucleoprotein 27kDa (U4/U6.U5) (212440_at), score: 0.68 SOAT1sterol O-acyltransferase 1 (221561_at), score: 0.71 SOLHsmall optic lobes homolog (Drosophila) (204275_at), score: -0.49 SORBS3sorbin and SH3 domain containing 3 (209253_at), score: -0.56 SP3Sp3 transcription factor (213168_at), score: 0.75 SPASTspastin (209748_at), score: 0.63 SPATA20spermatogenesis associated 20 (218164_at), score: -0.47 SRP19signal recognition particle 19kDa (205335_s_at), score: 0.69 SS18L1synovial sarcoma translocation gene on chromosome 18-like 1 (213140_s_at), score: 0.64 ST20suppressor of tumorigenicity 20 (210242_x_at), score: 0.67 STAT6signal transducer and activator of transcription 6, interleukin-4 induced (201331_s_at), score: -0.46 STRN3striatin, calmodulin binding protein 3 (204496_at), score: 0.63 STXBP3syntaxin binding protein 3 (203310_at), score: 0.72 SYNJ1synaptojanin 1 (212990_at), score: 0.75 TAF2TAF2 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 150kDa (209523_at), score: 0.88 TASP1taspase, threonine aspartase, 1 (219443_at), score: 0.64 TBCCD1TBCC domain containing 1 (206451_at), score: 0.68 TCEB1transcription elongation factor B (SIII), polypeptide 1 (15kDa, elongin C) (202823_at), score: 0.79 TCERG1transcription elongation regulator 1 (202396_at), score: 0.72 TDGthymine-DNA glycosylase (203743_s_at), score: 0.67 TENC1tensin like C1 domain containing phosphatase (tensin 2) (212494_at), score: -0.59 TGDSTDP-glucose 4,6-dehydratase (208249_s_at), score: 0.8 THUMPD1THUMP domain containing 1 (206555_s_at), score: 0.63 TIAF1TGFB1-induced anti-apoptotic factor 1 (202039_at), score: -0.48 TICAM2toll-like receptor adaptor molecule 2 (214658_at), score: 1 TM9SF3transmembrane 9 superfamily member 3 (217758_s_at), score: 0.79 TMBIM4transmembrane BAX inhibitor motif containing 4 (219206_x_at), score: 0.65 TMED7transmembrane emp24 protein transport domain containing 7 (209404_s_at), score: 0.69 TMEM123transmembrane protein 123 (211967_at), score: 0.74 TMEM30Atransmembrane protein 30A (217743_s_at), score: 0.67 TMF1TATA element modulatory factor 1 (213024_at), score: 0.77 TOMM20translocase of outer mitochondrial membrane 20 homolog (yeast) (200662_s_at), score: 0.72 TOPORStopoisomerase I binding, arginine/serine-rich (204071_s_at), score: 0.62 TSNAXtranslin-associated factor X (203983_at), score: 0.72 TXNDC9thioredoxin domain containing 9 (203008_x_at), score: 0.63 UBA2ubiquitin-like modifier activating enzyme 2 (201177_s_at), score: 0.72 UBE2V2ubiquitin-conjugating enzyme E2 variant 2 (209096_at), score: 0.71 UBE2Wubiquitin-conjugating enzyme E2W (putative) (218521_s_at), score: 0.65 UFM1ubiquitin-fold modifier 1 (218050_at), score: 0.74 UHRF1BP1LUHRF1 binding protein 1-like (213118_at), score: 0.74 USP46ubiquitin specific peptidase 46 (203869_at), score: 0.67 VEGFBvascular endothelial growth factor B (203683_s_at), score: -0.6 VENTXP1VENT homeobox (Xenopus laevis) pseudogene 1 (216722_at), score: -0.46 VPS13Cvacuolar protein sorting 13 homolog C (S. cerevisiae) (218396_at), score: 0.66 VPS37Bvacuolar protein sorting 37 homolog B (S. cerevisiae) (221704_s_at), score: -0.52 WBP2WW domain binding protein 2 (209117_at), score: -0.49 WDR4WD repeat domain 4 (221632_s_at), score: 0.61 WDR44WD repeat domain 44 (219297_at), score: 0.62 WRNWerner syndrome (205667_at), score: 0.62 WWTR1WW domain containing transcription regulator 1 (202132_at), score: 0.63 YEATS4YEATS domain containing 4 (218911_at), score: 0.63 YTHDC2YTH domain containing 2 (213077_at), score: 0.77 YTHDF3YTH domain family, member 3 (221749_at), score: 0.69 ZC3H14zinc finger CCCH-type containing 14 (213064_at), score: 0.77 ZCCHC10zinc finger, CCHC domain containing 10 (221193_s_at), score: 0.82 ZFAND1zinc finger, AN1-type domain 1 (218919_at), score: 0.64 ZFP36L1zinc finger protein 36, C3H type-like 1 (211965_at), score: -0.57 ZNF23zinc finger protein 23 (KOX 16) (213934_s_at), score: 0.67 ZNF267zinc finger protein 267 (219540_at), score: 0.63 ZNF580zinc finger protein 580 (220748_s_at), score: -0.48
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-TABM-263-raw-cel-1515486211.cel | 29 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| 10590_WBS.CEL | 2 | 8 | WBS | hgu133plus2 | WBS | WBS 1 |
| 10358_WBS.CEL | 1 | 8 | WBS | hgu133plus2 | WBS | WBS 1 |
| E-TABM-263-raw-cel-1515485691.cel | 3 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| ctrl a 08-03.CEL | 1 | 1 | DS-CC | hgu133a | none | DS-CC 1 |
| t21b 08-03.CEL | 5 | 1 | DS-CC | hgu133a | Down | DS-CC 5 |
| E-TABM-263-raw-cel-1515485871.cel | 12 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-GEOD-3860-raw-cel-1561690304.cel | 8 | 5 | HGPS | hgu133a | none | GMO8398C |
| E-TABM-263-raw-cel-1515486291.cel | 33 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485831.cel | 10 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| t21a 08-03.CEL | 4 | 1 | DS-CC | hgu133a | Down | DS-CC 4 |
| E-GEOD-3860-raw-cel-1561690376.cel | 13 | 5 | HGPS | hgu133a | HGPS | AG11513 |
| E-GEOD-3860-raw-cel-1561690272.cel | 7 | 5 | HGPS | hgu133a | HGPS | AG11498 |
| E-GEOD-4219-raw-cel-1311956275.cel | 8 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-TABM-263-raw-cel-1515485651.cel | 1 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-GEOD-4219-raw-cel-1311956398.cel | 12 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-TABM-263-raw-cel-1515485751.cel | 6 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486091.cel | 23 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485771.cel | 7 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485811.cel | 9 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486371.cel | 37 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485891.cel | 13 | 6 | Cycle | hgu133a2 | none | Cycle 1 |