
Under-expression is coded with green,
over-expression with red color.



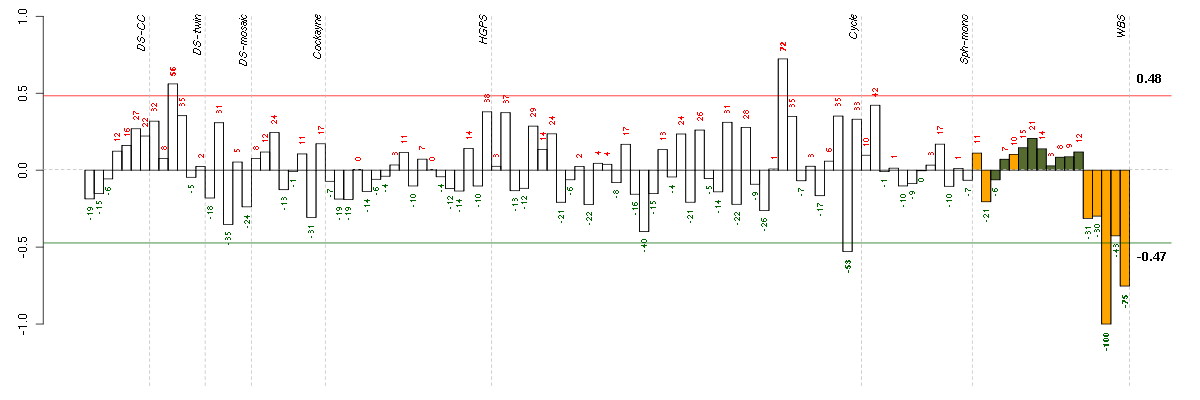
ATF4activating transcription factor 4 (tax-responsive enhancer element B67) (200779_at), score: 0.72 BET1Lblocked early in transport 1 homolog (S. cerevisiae)-like (220470_at), score: -0.64 C7orf58chromosome 7 open reading frame 58 (220032_at), score: -0.67 CHAC1ChaC, cation transport regulator homolog 1 (E. coli) (219270_at), score: 0.75 CHST7carbohydrate (N-acetylglucosamine 6-O) sulfotransferase 7 (206756_at), score: -0.63 DDIT3DNA-damage-inducible transcript 3 (209383_at), score: 1 DDIT4DNA-damage-inducible transcript 4 (202887_s_at), score: 0.78 HLXH2.0-like homeobox (214438_at), score: -0.67 IL27RAinterleukin 27 receptor, alpha (205926_at), score: -0.71 MAP2K3mitogen-activated protein kinase kinase 3 (215498_s_at), score: -0.64 MICAL3microtubule associated monoxygenase, calponin and LIM domain containing 3 (212715_s_at), score: -0.66 PGPEP1pyroglutamyl-peptidase I (219891_at), score: 0.78 TRIB3tribbles homolog 3 (Drosophila) (218145_at), score: 0.72
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| F223_WBS.CEL | 15 | 8 | WBS | hgu133plus2 | WBS | WBS 1 |
| H652_WBS.CEL | 17 | 8 | WBS | hgu133plus2 | WBS | WBS 1 |
| E-TABM-263-raw-cel-1515486411.cel | 39 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| 3Twin.CEL | 3 | 2 | DS-twin | hgu133plus2 | Down | DS-twin 3 |
| E-TABM-263-raw-cel-1515486271.cel | 32 | 6 | Cycle | hgu133a2 | none | Cycle 1 |