

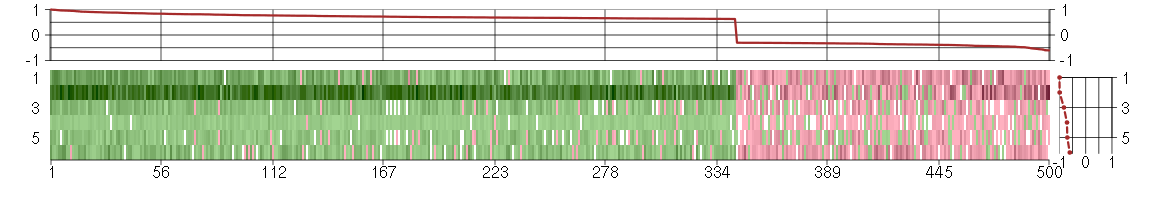

Under-expression is coded with green,
over-expression with red color.



ABI1abl-interactor 1 (209028_s_at), score: 0.84 ABOABO blood group (transferase A, alpha 1-3-N-acetylgalactosaminyltransferase; transferase B, alpha 1-3-galactosyltransferase) (216716_at), score: -0.31 ACADMacyl-Coenzyme A dehydrogenase, C-4 to C-12 straight chain (202502_at), score: 0.72 ACAP2ArfGAP with coiled-coil, ankyrin repeat and PH domains 2 (212476_at), score: 0.82 ACCN2amiloride-sensitive cation channel 2, neuronal (205156_s_at), score: -0.6 ACTR6ARP6 actin-related protein 6 homolog (yeast) (218395_at), score: 0.76 ADNPactivity-dependent neuroprotector homeobox (201773_at), score: 0.79 ADRA2Aadrenergic, alpha-2A-, receptor (209869_at), score: -0.31 AFF1AF4/FMR2 family, member 1 (201924_at), score: 0.66 AGLamylo-1, 6-glucosidase, 4-alpha-glucanotransferase (203566_s_at), score: 0.71 AKAP11A kinase (PRKA) anchor protein 11 (203156_at), score: 0.81 AKAP9A kinase (PRKA) anchor protein (yotiao) 9 (210962_s_at), score: 0.69 ANAPC13anaphase promoting complex subunit 13 (209001_s_at), score: 0.64 ANKRA2ankyrin repeat, family A (RFXANK-like), 2 (218769_s_at), score: 0.64 ANXA8L2annexin A8-like 2 (203074_at), score: -0.31 APOL1apolipoprotein L, 1 (209546_s_at), score: -0.32 APPBP2amyloid beta precursor protein (cytoplasmic tail) binding protein 2 (202629_at), score: 0.77 AQRaquarius homolog (mouse) (212584_at), score: 0.69 ARFGEF2ADP-ribosylation factor guanine nucleotide-exchange factor 2 (brefeldin A-inhibited) (218098_at), score: 0.95 ARFIP1ADP-ribosylation factor interacting protein 1 (218230_at), score: 0.81 ARGLU1arginine and glutamate rich 1 (218067_s_at), score: 0.67 ARHGAP12Rho GTPase activating protein 12 (207606_s_at), score: 0.65 ARHGEF12Rho guanine nucleotide exchange factor (GEF) 12 (201334_s_at), score: 0.95 ARID4AAT rich interactive domain 4A (RBP1-like) (205062_x_at), score: 0.72 ARID4BAT rich interactive domain 4B (RBP1-like) (221230_s_at), score: 0.63 ARL8BADP-ribosylation factor-like 8B (217852_s_at), score: 0.68 ARMC1armadillo repeat containing 1 (218185_s_at), score: 0.71 ARMC8armadillo repeat containing 8 (203486_s_at), score: 0.71 ASB9ankyrin repeat and SOCS box-containing 9 (205673_s_at), score: -0.53 ASNSD1asparagine synthetase domain containing 1 (217987_at), score: 0.73 ASPHD1aspartate beta-hydroxylase domain containing 1 (214993_at), score: -0.31 ASXL2additional sex combs like 2 (Drosophila) (218659_at), score: 0.73 ATF1activating transcription factor 1 (222103_at), score: 0.9 ATF2activating transcription factor 2 (212984_at), score: 0.82 ATG12ATG12 autophagy related 12 homolog (S. cerevisiae) (213026_at), score: 0.77 ATN1atrophin 1 (40489_at), score: -0.37 ATP11BATPase, class VI, type 11B (212536_at), score: 0.69 ATP2C1ATPase, Ca++ transporting, type 2C, member 1 (211137_s_at), score: 0.81 ATXN7ataxin 7 (204516_at), score: 0.66 AVL9AVL9 homolog (S. cerevisiase) (212474_at), score: 0.68 AZGP1alpha-2-glycoprotein 1, zinc-binding (217014_s_at), score: -0.3 B4GALNT1beta-1,4-N-acetyl-galactosaminyl transferase 1 (206435_at), score: -0.37 B4GALT1UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 1 (201883_s_at), score: -0.31 BACH1BTB and CNC homology 1, basic leucine zipper transcription factor 1 (204194_at), score: 0.66 BBS10Bardet-Biedl syndrome 10 (219487_at), score: 0.85 BCAS2breast carcinoma amplified sequence 2 (203053_at), score: 0.65 BMI1BMI1 polycomb ring finger oncogene (202265_at), score: 0.69 BMPR1Abone morphogenetic protein receptor, type IA (204832_s_at), score: 0.7 BNIP2BCL2/adenovirus E1B 19kDa interacting protein 2 (209308_s_at), score: 0.87 BRS3bombesin-like receptor 3 (207369_at), score: -0.33 BTAF1BTAF1 RNA polymerase II, B-TFIID transcription factor-associated, 170kDa (Mot1 homolog, S. cerevisiae) (209430_at), score: 0.64 BTBD1BTB (POZ) domain containing 1 (217945_at), score: 0.7 BZW1basic leucine zipper and W2 domains 1 (200777_s_at), score: 0.76 C10orf97chromosome 10 open reading frame 97 (218297_at), score: 0.77 C12orf29chromosome 12 open reading frame 29 (213701_at), score: 0.64 C14orf135chromosome 14 open reading frame 135 (219972_s_at), score: 0.75 C14orf147chromosome 14 open reading frame 147 (212460_at), score: 0.68 C16orf57chromosome 16 open reading frame 57 (218060_s_at), score: -0.37 C1DC1D nuclear receptor co-repressor (200056_s_at), score: 0.7 C1orf103chromosome 1 open reading frame 103 (220235_s_at), score: 0.72 C1orf63chromosome 1 open reading frame 63 (209006_s_at), score: 0.67 C20orf12chromosome 20 open reading frame 12 (219951_s_at), score: -0.3 C2orf72chromosome 2 open reading frame 72 (213143_at), score: -0.34 C3orf63chromosome 3 open reading frame 63 (209284_s_at), score: 0.7 C4orf16chromosome 4 open reading frame 16 (219023_at), score: 0.66 C5orf22chromosome 5 open reading frame 22 (203738_at), score: 0.77 C5orf28chromosome 5 open reading frame 28 (219029_at), score: 0.97 C6orf211chromosome 6 open reading frame 211 (218195_at), score: 0.86 C6orf47chromosome 6 open reading frame 47 (204968_at), score: -0.31 C7orf28Achromosome 7 open reading frame 28A (201974_s_at), score: -0.45 C9orf82chromosome 9 open reading frame 82 (219276_x_at), score: 0.76 CAB39calcium binding protein 39 (217873_at), score: 0.7 CAPZA2capping protein (actin filament) muscle Z-line, alpha 2 (201237_at), score: 0.89 CASP3caspase 3, apoptosis-related cysteine peptidase (202763_at), score: 0.7 CCNCcyclin C (201955_at), score: 0.63 CCNE1cyclin E1 (213523_at), score: -0.31 CCNT2cyclin T2 (204645_at), score: 0.7 CD2APCD2-associated protein (203593_at), score: 0.73 CD83CD83 molecule (204440_at), score: -0.33 CDC37L1cell division cycle 37 homolog (S. cerevisiae)-like 1 (219343_at), score: 0.64 CDC73cell division cycle 73, Paf1/RNA polymerase II complex component, homolog (S. cerevisiae) (218578_at), score: 0.76 CDKN2AIPCDKN2A interacting protein (218929_at), score: 0.65 CELSR3cadherin, EGF LAG seven-pass G-type receptor 3 (flamingo homolog, Drosophila) (40020_at), score: -0.43 CEP170centrosomal protein 170kDa (212746_s_at), score: 0.77 CHMP5chromatin modifying protein 5 (218085_at), score: 0.75 CIDECcell death-inducing DFFA-like effector c (219398_at), score: -0.31 CLCN3chloride channel 3 (201734_at), score: 0.67 CLDND1claudin domain containing 1 (208925_at), score: 0.69 CLEC7AC-type lectin domain family 7, member A (221698_s_at), score: -0.36 CLIP1CAP-GLY domain containing linker protein 1 (210716_s_at), score: 0.76 CLK1CDC-like kinase 1 (214683_s_at), score: 0.67 CLPXClpX caseinolytic peptidase X homolog (E. coli) (204809_at), score: 0.68 CMPK1cytidine monophosphate (UMP-CMP) kinase 1, cytosolic (217870_s_at), score: 0.8 CMTM6CKLF-like MARVEL transmembrane domain containing 6 (217947_at), score: 0.67 COL7A1collagen, type VII, alpha 1 (217312_s_at), score: -0.31 COMMD8COMM domain containing 8 (218351_at), score: 0.72 COPB1coatomer protein complex, subunit beta 1 (201359_at), score: 0.64 COPS2COP9 constitutive photomorphogenic homolog subunit 2 (Arabidopsis) (202467_s_at), score: 0.66 CORO2Acoronin, actin binding protein, 2A (205538_at), score: -0.33 COX11COX11 homolog, cytochrome c oxidase assembly protein (yeast) (211727_s_at), score: 0.7 COX6A2cytochrome c oxidase subunit VIa polypeptide 2 (206353_at), score: -0.45 CREB1cAMP responsive element binding protein 1 (204314_s_at), score: 0.7 CREBBPCREB binding protein (202160_at), score: 0.67 CSGALNACT2chondroitin sulfate N-acetylgalactosaminyltransferase 2 (222235_s_at), score: 0.66 CSNK1G3casein kinase 1, gamma 3 (220768_s_at), score: 0.88 CTR9Ctr9, Paf1/RNA polymerase II complex component, homolog (S. cerevisiae) (202060_at), score: 0.64 CTSDcathepsin D (200766_at), score: -0.54 CUL5cullin 5 (203531_at), score: 0.87 CYB5R4cytochrome b5 reductase 4 (219079_at), score: 0.64 CYP2A6cytochrome P450, family 2, subfamily A, polypeptide 6 (211295_x_at), score: -0.32 DCUN1D1DCN1, defective in cullin neddylation 1, domain containing 1 (S. cerevisiae) (218583_s_at), score: 0.81 DCUN1D4DCN1, defective in cullin neddylation 1, domain containing 4 (S. cerevisiae) (212855_at), score: 0.78 DEKDEK oncogene (200934_at), score: 0.74 DHX29DEAH (Asp-Glu-Ala-His) box polypeptide 29 (212648_at), score: 0.8 DLATdihydrolipoamide S-acetyltransferase (212568_s_at), score: 0.65 DLDdihydrolipoamide dehydrogenase (209095_at), score: 0.77 DMXL1Dmx-like 1 (203791_at), score: 0.67 DNAJB14DnaJ (Hsp40) homolog, subfamily B, member 14 (219237_s_at), score: 0.8 DNAJC10DnaJ (Hsp40) homolog, subfamily C, member 10 (221781_s_at), score: 0.63 DNAJC13DnaJ (Hsp40) homolog, subfamily C, member 13 (212467_at), score: 0.65 DNAJC25-GNG10DNAJC25-GNG10 readthrough transcript (201921_at), score: 0.79 DOCK6dedicator of cytokinesis 6 (221794_at), score: -0.41 DOHHdeoxyhypusine hydroxylase/monooxygenase (208141_s_at), score: -0.41 DPY19L4dpy-19-like 4 (C. elegans) (213391_at), score: 0.71 EAPPE2F-associated phosphoprotein (202623_at), score: 0.73 ECHDC1enoyl Coenzyme A hydratase domain containing 1 (219974_x_at), score: 0.76 EDEM3ER degradation enhancer, mannosidase alpha-like 3 (220342_x_at), score: 0.64 EFHA1EF-hand domain family, member A1 (212410_at), score: 0.81 EGR3early growth response 3 (206115_at), score: -0.31 EIF2AK3eukaryotic translation initiation factor 2-alpha kinase 3 (218696_at), score: 0.68 EIF4G2eukaryotic translation initiation factor 4 gamma, 2 (200004_at), score: 0.66 EML5echinoderm microtubule associated protein like 5 (213063_at), score: 0.67 EP300E1A binding protein p300 (202221_s_at), score: 0.68 EPM2AIP1EPM2A (laforin) interacting protein 1 (202909_at), score: 0.66 EPN1epsin 1 (221141_x_at), score: -0.32 EPOerythropoietin (217254_s_at), score: -0.34 EPS15epidermal growth factor receptor pathway substrate 15 (217887_s_at), score: 0.88 EPS8epidermal growth factor receptor pathway substrate 8 (202609_at), score: 0.68 ERBB2IPerbb2 interacting protein (217941_s_at), score: 0.79 ETNK2ethanolamine kinase 2 (219268_at), score: -0.45 FAM18Bfamily with sequence similarity 18, member B (218446_s_at), score: 0.78 FAM21Cfamily with sequence similarity 21, member C (211068_x_at), score: 0.65 FAM3Cfamily with sequence similarity 3, member C (201889_at), score: 0.63 FAM53Bfamily with sequence similarity 53, member B (203206_at), score: -0.37 FAM8A1family with sequence similarity 8, member A1 (203420_at), score: 0.63 FBRSfibrosin (218255_s_at), score: -0.34 FBXL5F-box and leucine-rich repeat protein 5 (209004_s_at), score: 0.71 FBXO3F-box protein 3 (218432_at), score: 0.96 FBXW7F-box and WD repeat domain containing 7 (218751_s_at), score: -0.52 FGF1fibroblast growth factor 1 (acidic) (205117_at), score: -0.36 FKBP4FK506 binding protein 4, 59kDa (200895_s_at), score: -0.38 FMR1fragile X mental retardation 1 (215245_x_at), score: 0.85 FOSBFBJ murine osteosarcoma viral oncogene homolog B (202768_at), score: -0.6 FOSL1FOS-like antigen 1 (204420_at), score: -0.32 FOXO3Bforkhead box O3B pseudogene (210655_s_at), score: -0.43 G3BP2GTPase activating protein (SH3 domain) binding protein 2 (208841_s_at), score: 0.65 GAB1GRB2-associated binding protein 1 (207112_s_at), score: -0.33 GBA3glucosidase, beta, acid 3 (cytosolic) (219954_s_at), score: -0.32 GBASglioblastoma amplified sequence (201816_s_at), score: 0.66 GCC2GRIP and coiled-coil domain containing 2 (202832_at), score: 0.88 GFOD2glucose-fructose oxidoreductase domain containing 2 (221028_s_at), score: -0.31 GMFBglia maturation factor, beta (202543_s_at), score: 0.7 GNAI1guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 1 (209576_at), score: 0.72 GNAQguanine nucleotide binding protein (G protein), q polypeptide (202615_at), score: 0.7 GNG12guanine nucleotide binding protein (G protein), gamma 12 (212294_at), score: 0.73 GOLGA4golgi autoantigen, golgin subfamily a, 4 (201567_s_at), score: 0.76 GOLGA7golgi autoantigen, golgin subfamily a, 7 (217819_at), score: 0.64 GPR63G protein-coupled receptor 63 (220993_s_at), score: -0.32 GRINL1Aglutamate receptor, ionotropic, N-methyl D-aspartate-like 1A (212244_at), score: 0.74 GSK3Aglycogen synthase kinase 3 alpha (632_at), score: -0.35 HAB1B1 for mucin (215778_x_at), score: -0.5 HBEGFheparin-binding EGF-like growth factor (203821_at), score: -0.46 HDAC2histone deacetylase 2 (201833_at), score: 0.71 HIF1Ahypoxia inducible factor 1, alpha subunit (basic helix-loop-helix transcription factor) (200989_at), score: 0.69 HIGD1AHIG1 domain family, member 1A (221896_s_at), score: 0.77 HISPPD1histidine acid phosphatase domain containing 1 (203253_s_at), score: 0.98 HIST1H2ALhistone cluster 1, H2al (214554_at), score: -0.32 HMGN4high mobility group nucleosomal binding domain 4 (209786_at), score: 0.7 HNRNPH2heterogeneous nuclear ribonucleoprotein H2 (H') (201132_at), score: 0.83 HOMER2homer homolog 2 (Drosophila) (217080_s_at), score: -0.3 HOOK1hook homolog 1 (Drosophila) (219976_at), score: -0.39 HOXA4homeobox A4 (206289_at), score: -0.32 HSD17B11hydroxysteroid (17-beta) dehydrogenase 11 (217989_at), score: 0.73 HSD17B12hydroxysteroid (17-beta) dehydrogenase 12 (217869_at), score: 0.64 HSPA6heat shock 70kDa protein 6 (HSP70B') (117_at), score: -0.41 HSPB6heat shock protein, alpha-crystallin-related, B6 (214767_s_at), score: -0.34 HUNKhormonally up-regulated Neu-associated kinase (219535_at), score: -0.44 IBTKinhibitor of Bruton agammaglobulinemia tyrosine kinase (210970_s_at), score: 0.77 IL16interleukin 16 (lymphocyte chemoattractant factor) (209827_s_at), score: -0.43 IMPA1inositol(myo)-1(or 4)-monophosphatase 1 (203011_at), score: 0.77 IMPACTImpact homolog (mouse) (218637_at), score: 0.78 ITGB3integrin, beta 3 (platelet glycoprotein IIIa, antigen CD61) (204628_s_at), score: -0.43 ITPKCinositol 1,4,5-trisphosphate 3-kinase C (213076_at), score: -0.39 ITSN2intersectin 2 (209898_x_at), score: 0.86 JMJD1Cjumonji domain containing 1C (221763_at), score: 0.86 JMJD3jumonji domain containing 3, histone lysine demethylase (213146_at), score: -0.43 JUNDjun D proto-oncogene (203751_x_at), score: -0.5 KCNQ3potassium voltage-gated channel, KQT-like subfamily, member 3 (206573_at), score: -0.34 KCTD17potassium channel tetramerisation domain containing 17 (205561_at), score: -0.38 KCTD9potassium channel tetramerisation domain containing 9 (218823_s_at), score: 0.84 KIAA0196KIAA0196 (201985_at), score: 0.74 KIAA0528KIAA0528 (212943_at), score: 0.65 KIAA1012KIAA1012 (207305_s_at), score: 0.75 KIAA1033KIAA1033 (215936_s_at), score: 0.63 KIAA1109KIAA1109 (212779_at), score: 0.9 KIAA1128KIAA1128 (209379_s_at), score: 0.73 KIAA1324KIAA1324 (221874_at), score: -0.37 KIR3DX1killer cell immunoglobulin-like receptor, three domains, X1 (216428_x_at), score: -0.36 KRCC1lysine-rich coiled-coil 1 (218303_x_at), score: 0.67 KRT8P12keratin 8 pseudogene 12 (222060_at), score: -0.39 LAMB4laminin, beta 4 (215516_at), score: -0.4 LANCL1LanC lantibiotic synthetase component C-like 1 (bacterial) (202020_s_at), score: 0.7 LAPTM5lysosomal multispanning membrane protein 5 (201721_s_at), score: -0.34 LARP7La ribonucleoprotein domain family, member 7 (212785_s_at), score: 0.93 LMCD1LIM and cysteine-rich domains 1 (218574_s_at), score: -0.32 LOC100129502hypothetical protein LOC100129502 (217548_at), score: -0.31 LOC100132072hypothetical protein LOC100132072 (220175_s_at), score: 0.71 LOC220594TL132 protein (213510_x_at), score: 0.68 LOC286434hypothetical protein LOC286434 (222196_at), score: -0.31 LOC388152hypothetical LOC388152 (220602_s_at), score: -0.31 LOC440345hypothetical protein LOC440345 (214984_at), score: -0.33 LOH3CR2Aloss of heterozygosity, 3, chromosomal region 2, gene A (220244_at), score: -0.44 LONRF3LON peptidase N-terminal domain and ring finger 3 (220009_at), score: -0.34 LPAR4lysophosphatidic acid receptor 4 (206960_at), score: -0.46 LPGAT1lysophosphatidylglycerol acyltransferase 1 (202651_at), score: 0.84 LRP1low density lipoprotein-related protein 1 (alpha-2-macroglobulin receptor) (200784_s_at), score: -0.31 LRRC40leucine rich repeat containing 40 (218577_at), score: 0.89 LRRN2leucine rich repeat neuronal 2 (216164_at), score: -0.38 LSM14ALSM14A, SCD6 homolog A (S. cerevisiae) (212131_at), score: 0.71 LUC7L2LUC7-like 2 (S. cerevisiae) (220099_s_at), score: 0.7 LZTFL1leucine zipper transcription factor-like 1 (218437_s_at), score: 0.68 MAP1Smicrotubule-associated protein 1S (218522_s_at), score: -0.33 MAP2K3mitogen-activated protein kinase kinase 3 (215498_s_at), score: -0.37 MAP4K3mitogen-activated protein kinase kinase kinase kinase 3 (218311_at), score: 0.87 MAPK6mitogen-activated protein kinase 6 (207121_s_at), score: 0.78 MAPK8IP1mitogen-activated protein kinase 8 interacting protein 1 (213013_at), score: -0.48 MAPKSP1MAPK scaffold protein 1 (217971_at), score: 0.86 MAPRE3microtubule-associated protein, RP/EB family, member 3 (203842_s_at), score: -0.33 MAT2Bmethionine adenosyltransferase II, beta (217993_s_at), score: 0.71 MATR3matrin 3 (200626_s_at), score: 0.84 MBIPMAP3K12 binding inhibitory protein 1 (218411_s_at), score: 0.81 MBNL1muscleblind-like (Drosophila) (201151_s_at), score: 0.76 MC1Rmelanocortin 1 receptor (alpha melanocyte stimulating hormone receptor) (205458_at), score: -0.43 MECRmitochondrial trans-2-enoyl-CoA reductase (218664_at), score: -0.32 MED13mediator complex subunit 13 (201986_at), score: 0.71 MED23mediator complex subunit 23 (218846_at), score: 0.8 MED9mediator complex subunit 9 (218372_at), score: -0.41 MEX3Cmex-3 homolog C (C. elegans) (218247_s_at), score: 0.69 MFFmitochondrial fission factor (219137_s_at), score: 0.64 MFN1mitofusin 1 (217043_s_at), score: 0.63 MICAL3microtubule associated monoxygenase, calponin and LIM domain containing 3 (212715_s_at), score: -0.37 MMADHCmethylmalonic aciduria (cobalamin deficiency) cblD type, with homocystinuria (217883_at), score: 0.63 MMRN2multimerin 2 (219091_s_at), score: -0.33 MORC3MORC family CW-type zinc finger 3 (213000_at), score: 0.73 MORF4mortality factor 4 (221381_s_at), score: 0.67 MORF4L1mortality factor 4 like 1 (217982_s_at), score: 0.72 MOSPD1motile sperm domain containing 1 (218853_s_at), score: 0.66 MPP2membrane protein, palmitoylated 2 (MAGUK p55 subfamily member 2) (213270_at), score: -0.37 MSTP9macrophage stimulating, pseudogene 9 (213382_at), score: -0.34 MTMR6myotubularin related protein 6 (214429_at), score: 0.69 MUC1mucin 1, cell surface associated (207847_s_at), score: -0.32 MUC5ACmucin 5AC, oligomeric mucus/gel-forming (217182_at), score: -0.42 MYCBP2MYC binding protein 2 (201960_s_at), score: 0.7 MYO1Bmyosin IB (212364_at), score: 0.64 MYO6myosin VI (203216_s_at), score: 0.82 N4BP3Nedd4 binding protein 3 (214775_at), score: -0.33 NCAPH2non-SMC condensin II complex, subunit H2 (40640_at), score: -0.31 NCK1NCK adaptor protein 1 (211063_s_at), score: 0.83 NDUFA5NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 5, 13kDa (201304_at), score: 0.83 NEUROD2neurogenic differentiation 2 (210271_at), score: -0.31 NFATC1nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 1 (210162_s_at), score: -0.42 NPnucleoside phosphorylase (201695_s_at), score: -0.31 NPHP4nephronophthisis 4 (213471_at), score: -0.36 NPTNneuroplastin (202228_s_at), score: 0.63 NRN1neuritin 1 (218625_at), score: -0.43 NT5C25'-nucleotidase, cytosolic II (209155_s_at), score: 0.79 NXT1NTF2-like export factor 1 (218708_at), score: -0.32 OATornithine aminotransferase (gyrate atrophy) (201599_at), score: 0.74 OGFOD22-oxoglutarate and iron-dependent oxygenase domain containing 2 (219246_s_at), score: -0.37 OGTO-linked N-acetylglucosamine (GlcNAc) transferase (UDP-N-acetylglucosamine:polypeptide-N-acetylglucosaminyl transferase) (209240_at), score: 0.73 OPA1optic atrophy 1 (autosomal dominant) (212213_x_at), score: 0.9 OR1E2olfactory receptor, family 1, subfamily E, member 2 (208587_s_at), score: -0.57 OSBPL8oxysterol binding protein-like 8 (212582_at), score: 0.75 PAQR3progestin and adipoQ receptor family member III (213372_at), score: 0.64 PBRM1polybromo 1 (220355_s_at), score: 0.73 PCLOpiccolo (presynaptic cytomatrix protein) (213558_at), score: -0.34 PCMTD2protein-L-isoaspartate (D-aspartate) O-methyltransferase domain containing 2 (212406_s_at), score: 0.67 PCNPPEST proteolytic signal containing nuclear protein (217816_s_at), score: 0.88 PDCD10programmed cell death 10 (210907_s_at), score: 0.73 PDK2pyruvate dehydrogenase kinase, isozyme 2 (202590_s_at), score: -0.32 PDS5APDS5, regulator of cohesion maintenance, homolog A (S. cerevisiae) (212138_at), score: 0.78 PDS5BPDS5, regulator of cohesion maintenance, homolog B (S. cerevisiae) (207956_x_at), score: 0.69 PEX6peroxisomal biogenesis factor 6 (204545_at), score: -0.36 PHF3PHD finger protein 3 (217954_s_at), score: 0.74 PIK3C2Aphosphoinositide-3-kinase, class 2, alpha polypeptide (213070_at), score: 0.82 PIK3CAphosphoinositide-3-kinase, catalytic, alpha polypeptide (204369_at), score: 0.73 PIP5K1Cphosphatidylinositol-4-phosphate 5-kinase, type I, gamma (212518_at), score: -0.35 PIP5K3phosphatidylinositol-3-phosphate/phosphatidylinositol 5-kinase, type III (213111_at), score: 0.75 PJA2praja ring finger 2 (201133_s_at), score: 1 PKLRpyruvate kinase, liver and RBC (222078_at), score: -0.37 PLAURplasminogen activator, urokinase receptor (211924_s_at), score: -0.38 PLEKHG6pleckstrin homology domain containing, family G (with RhoGef domain) member 6 (220073_s_at), score: -0.36 PMEPA1prostate transmembrane protein, androgen induced 1 (217875_s_at), score: -0.48 PMS1PMS1 postmeiotic segregation increased 1 (S. cerevisiae) (213677_s_at), score: 0.69 PMS2L2postmeiotic segregation increased 2-like 2 pseudogene (215410_at), score: -0.37 PNPLA2patatin-like phospholipase domain containing 2 (212705_x_at), score: -0.56 POLR2Bpolymerase (RNA) II (DNA directed) polypeptide B, 140kDa (201803_at), score: 0.7 PPFIA1protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 1 (202066_at), score: 0.68 PPIAL4Apeptidylprolyl isomerase A (cyclophilin A)-like 4A (217136_at), score: -0.31 PPM1Bprotein phosphatase 1B (formerly 2C), magnesium-dependent, beta isoform (213225_at), score: 0.69 PPP1CCprotein phosphatase 1, catalytic subunit, gamma isoform (200726_at), score: 0.65 PPWD1peptidylprolyl isomerase domain and WD repeat containing 1 (213483_at), score: 0.65 PRB3proline-rich protein BstNI subfamily 3 (206998_x_at), score: -0.39 PRKRIRprotein-kinase, interferon-inducible double stranded RNA dependent inhibitor, repressor of (P58 repressor) (209323_at), score: 0.66 PRLRprolactin receptor (216638_s_at), score: -0.47 PRPF18PRP18 pre-mRNA processing factor 18 homolog (S. cerevisiae) (221547_at), score: 0.76 PRRG1proline rich Gla (G-carboxyglutamic acid) 1 (205618_at), score: 0.74 PRSS7protease, serine, 7 (enterokinase) (217269_s_at), score: -0.45 PTGDSprostaglandin D2 synthase 21kDa (brain) (212187_x_at), score: -0.31 PTP4A2protein tyrosine phosphatase type IVA, member 2 (208615_s_at), score: 0.68 PUM2pumilio homolog 2 (Drosophila) (216221_s_at), score: 0.95 PYROXD1pyridine nucleotide-disulphide oxidoreductase domain 1 (213878_at), score: 0.66 RAB1ARAB1A, member RAS oncogene family (213440_at), score: 0.69 RAB6ARAB6A, member RAS oncogene family (201047_x_at), score: 0.63 RANBP6RAN binding protein 6 (213019_at), score: 0.72 RAP1ARAP1A, member of RAS oncogene family (202362_at), score: 0.76 RAP1BRAP1B, member of RAS oncogene family (200833_s_at), score: 0.67 RAP2CRAP2C, member of RAS oncogene family (218668_s_at), score: 0.69 RB1retinoblastoma 1 (203132_at), score: 0.76 RB1CC1RB1-inducible coiled-coil 1 (202034_x_at), score: 0.79 RBM26RNA binding motif protein 26 (218422_s_at), score: 0.63 RCHY1ring finger and CHY zinc finger domain containing 1 (212749_s_at), score: 0.69 RCN2reticulocalbin 2, EF-hand calcium binding domain (201485_s_at), score: 0.87 RECQLRecQ protein-like (DNA helicase Q1-like) (205091_x_at), score: 0.88 REV1REV1 homolog (S. cerevisiae) (218428_s_at), score: 0.64 RFC1replication factor C (activator 1) 1, 145kDa (209085_x_at), score: 0.73 RGS3regulator of G-protein signaling 3 (203823_at), score: -0.44 RHOT1ras homolog gene family, member T1 (222148_s_at), score: 0.64 RIF1RAP1 interacting factor homolog (yeast) (214700_x_at), score: 0.77 RND3Rho family GTPase 3 (212724_at), score: 0.83 RNF111ring finger protein 111 (218761_at), score: 0.66 RNF160ring finger protein 160 (215596_s_at), score: 0.7 ROCK1Rho-associated, coiled-coil containing protein kinase 1 (213044_at), score: 0.81 ROCK2Rho-associated, coiled-coil containing protein kinase 2 (202762_at), score: 0.64 RP2retinitis pigmentosa 2 (X-linked recessive) (205191_at), score: 0.74 RPAINRPA interacting protein (216961_s_at), score: -0.31 RPAP3RNA polymerase II associated protein 3 (218842_at), score: 0.64 RPH3ALrabphilin 3A-like (without C2 domains) (221614_s_at), score: -0.48 RPL9ribosomal protein L9 (200032_s_at), score: 0.67 RPS13ribosomal protein S13 (200018_at), score: 0.68 RPS6KA1ribosomal protein S6 kinase, 90kDa, polypeptide 1 (203379_at), score: -0.4 RRADRas-related associated with diabetes (204803_s_at), score: -0.6 RSL24D1ribosomal L24 domain containing 1 (217915_s_at), score: 0.7 RSRC2arginine/serine-rich coiled-coil 2 (202301_s_at), score: 0.7 RUNX3runt-related transcription factor 3 (204198_s_at), score: -0.38 SACM1LSAC1 suppressor of actin mutations 1-like (yeast) (202797_at), score: 0.77 SC5DLsterol-C5-desaturase (ERG3 delta-5-desaturase homolog, S. cerevisiae)-like (211423_s_at), score: 0.65 SCFD1sec1 family domain containing 1 (215548_s_at), score: 0.75 SDCBPsyndecan binding protein (syntenin) (200958_s_at), score: 0.92 SDCCAG1serologically defined colon cancer antigen 1 (218649_x_at), score: 0.72 SEC22BSEC22 vesicle trafficking protein homolog B (S. cerevisiae) (214257_s_at), score: 0.68 SEC24BSEC24 family, member B (S. cerevisiae) (202798_at), score: 0.89 SEC24DSEC24 family, member D (S. cerevisiae) (202375_at), score: 0.65 SEC31ASEC31 homolog A (S. cerevisiae) (215009_s_at), score: 0.72 SENP6SUMO1/sentrin specific peptidase 6 (202318_s_at), score: 0.8 SEPT5septin 5 (209767_s_at), score: -0.44 SEPT7septin 7 (213151_s_at), score: 0.68 SERINC1serine incorporator 1 (208671_at), score: 0.85 SERPINA1serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 1 (211429_s_at), score: -0.38 SFRS12splicing factor, arginine/serine-rich 12 (212721_at), score: 0.79 SH3D19SH3 domain containing 19 (211620_x_at), score: -0.45 SHBSrc homology 2 domain containing adaptor protein B (204657_s_at), score: -0.37 SHOC2soc-2 suppressor of clear homolog (C. elegans) (202777_at), score: 0.85 SHPKsedoheptulokinase (219713_at), score: -0.32 SLC10A1solute carrier family 10 (sodium/bile acid cotransporter family), member 1 (207185_at), score: -0.33 SLC19A1solute carrier family 19 (folate transporter), member 1 (211576_s_at), score: -0.33 SLC1A2solute carrier family 1 (glial high affinity glutamate transporter), member 2 (208389_s_at), score: -0.33 SLC25A24solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 24 (204342_at), score: 0.9 SLC25A36solute carrier family 25, member 36 (201918_at), score: 0.65 SLC25A46solute carrier family 25, member 46 (212833_at), score: 0.64 SLC30A9solute carrier family 30 (zinc transporter), member 9 (202614_at), score: 0.67 SLC35A3solute carrier family 35 (UDP-N-acetylglucosamine (UDP-GlcNAc) transporter), member A3 (206770_s_at), score: 0.74 SLC38A2solute carrier family 38, member 2 (220924_s_at), score: 0.73 SLC9A6solute carrier family 9 (sodium/hydrogen exchanger), member 6 (203909_at), score: 0.73 SLKSTE20-like kinase (yeast) (206875_s_at), score: 0.99 SLMO2slowmo homolog 2 (Drosophila) (217851_s_at), score: 0.88 SLTMSAFB-like, transcription modulator (217828_at), score: 0.82 SMAD4SMAD family member 4 (202527_s_at), score: 0.76 SMCHD1structural maintenance of chromosomes flexible hinge domain containing 1 (212569_at), score: 0.66 SMNDC1survival motor neuron domain containing 1 (200071_at), score: 0.63 SNRNP27small nuclear ribonucleoprotein 27kDa (U4/U6.U5) (212440_at), score: 0.7 SNX13sorting nexin 13 (213292_s_at), score: 0.67 SNX2sorting nexin 2 (202113_s_at), score: 0.64 SOCS1suppressor of cytokine signaling 1 (210001_s_at), score: -0.36 SOCS3suppressor of cytokine signaling 3 (206359_at), score: -0.4 SOCS5suppressor of cytokine signaling 5 (209647_s_at), score: 0.67 SOS2son of sevenless homolog 2 (Drosophila) (212870_at), score: 0.74 SP3Sp3 transcription factor (213168_at), score: 0.74 SPANXCSPANX family, member C (220217_x_at), score: -0.34 SPASTspastin (209748_at), score: 0.69 SPATA5L1spermatogenesis associated 5-like 1 (222163_s_at), score: 0.68 SPG20spastic paraplegia 20 (Troyer syndrome) (212526_at), score: 0.8 SPSB1splA/ryanodine receptor domain and SOCS box containing 1 (219677_at), score: -0.33 SRP19signal recognition particle 19kDa (205335_s_at), score: 0.66 SRP54signal recognition particle 54kDa (203605_at), score: 0.73 SRP9signal recognition particle 9kDa (201273_s_at), score: 0.81 ST3GAL1ST3 beta-galactoside alpha-2,3-sialyltransferase 1 (208322_s_at), score: -0.32 STAM2signal transducing adaptor molecule (SH3 domain and ITAM motif) 2 (209649_at), score: 0.82 STARD3StAR-related lipid transfer (START) domain containing 3 (202991_at), score: -0.38 STRN3striatin, calmodulin binding protein 3 (204496_at), score: 0.94 STXBP3syntaxin binding protein 3 (203310_at), score: 0.91 SUMO1SMT3 suppressor of mif two 3 homolog 1 (S. cerevisiae) (208761_s_at), score: 0.84 SYNPOsynaptopodin (202796_at), score: -0.38 SYPL1synaptophysin-like 1 (201260_s_at), score: 0.7 TAF1BTATA box binding protein (TBP)-associated factor, RNA polymerase I, B, 63kDa (216941_s_at), score: 0.63 TAF2TAF2 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 150kDa (209523_at), score: 0.84 TAF4BTAF4b RNA polymerase II, TATA box binding protein (TBP)-associated factor, 105kDa (216226_at), score: -0.33 TANKTRAF family member-associated NFKB activator (207616_s_at), score: 0.65 TBC1D15TBC1 domain family, member 15 (218268_at), score: 0.78 TBK1TANK-binding kinase 1 (218520_at), score: 0.91 TCERG1transcription elongation regulator 1 (202396_at), score: 0.67 TCF12transcription factor 12 (208986_at), score: 0.75 TFB2Mtranscription factor B2, mitochondrial (218605_at), score: 0.65 TGFAtransforming growth factor, alpha (205016_at), score: -0.46 TGIF2TGFB-induced factor homeobox 2 (216262_s_at), score: -0.31 TGM4transglutaminase 4 (prostate) (217566_s_at), score: -0.47 THBDthrombomodulin (203887_s_at), score: -0.33 THUMPD1THUMP domain containing 1 (206555_s_at), score: 0.67 TIA1TIA1 cytotoxic granule-associated RNA binding protein (201446_s_at), score: 0.66 TICAM2toll-like receptor adaptor molecule 2 (214658_at), score: 0.78 TLK1tousled-like kinase 1 (202606_s_at), score: 0.81 TM9SF2transmembrane 9 superfamily member 2 (201078_at), score: 0.63 TM9SF3transmembrane 9 superfamily member 3 (217758_s_at), score: 0.71 TMBIM4transmembrane BAX inhibitor motif containing 4 (219206_x_at), score: 0.66 TMCC2transmembrane and coiled-coil domain family 2 (213096_at), score: -0.32 TMED7transmembrane emp24 protein transport domain containing 7 (209404_s_at), score: 0.91 TMEM123transmembrane protein 123 (211967_at), score: 0.96 TMEM149transmembrane protein 149 (219690_at), score: -0.39 TMEM30Atransmembrane protein 30A (217743_s_at), score: 0.91 TMEM50Atransmembrane protein 50A (217766_s_at), score: 0.67 TMF1TATA element modulatory factor 1 (213024_at), score: 0.99 TNFSF9tumor necrosis factor (ligand) superfamily, member 9 (206907_at), score: -0.43 TNKS2tankyrase, TRF1-interacting ankyrin-related ADP-ribose polymerase 2 (218228_s_at), score: 0.63 TOP2Btopoisomerase (DNA) II beta 180kDa (211987_at), score: 0.96 TOPORStopoisomerase I binding, arginine/serine-rich (204071_s_at), score: 0.84 TP53I11tumor protein p53 inducible protein 11 (214667_s_at), score: -0.39 TRAM1translocation associated membrane protein 1 (210733_at), score: 0.72 TRAPPC2trafficking protein particle complex 2 (219351_at), score: 0.7 TRMT11tRNA methyltransferase 11 homolog (S. cerevisiae) (218877_s_at), score: 0.79 TRPC3transient receptor potential cation channel, subfamily C, member 3 (210814_at), score: -0.56 TSNAXtranslin-associated factor X (203983_at), score: 0.84 TSPYL4TSPY-like 4 (212928_at), score: 0.65 TTC33tetratricopeptide repeat domain 33 (219421_at), score: 0.63 TTC37tetratricopeptide repeat domain 37 (203048_s_at), score: 0.69 TTRAPTRAF and TNF receptor associated protein (202266_at), score: 0.64 TXNDC9thioredoxin domain containing 9 (203008_x_at), score: 0.66 UBA3ubiquitin-like modifier activating enzyme 3 (209115_at), score: 0.81 UBE2Bubiquitin-conjugating enzyme E2B (RAD6 homolog) (211763_s_at), score: 0.67 UBE2D1ubiquitin-conjugating enzyme E2D 1 (UBC4/5 homolog, yeast) (211764_s_at), score: 0.67 UBE2E1ubiquitin-conjugating enzyme E2E 1 (UBC4/5 homolog, yeast) (212519_at), score: 0.69 UBE2J1ubiquitin-conjugating enzyme E2, J1 (UBC6 homolog, yeast) (217826_s_at), score: 0.73 UBE2V2ubiquitin-conjugating enzyme E2 variant 2 (209096_at), score: 0.66 UBXN7UBX domain protein 7 (212840_at), score: 0.68 UEVLDUEV and lactate/malate dehyrogenase domains (220775_s_at), score: 0.66 UFM1ubiquitin-fold modifier 1 (218050_at), score: 0.68 UNC50unc-50 homolog (C. elegans) (203583_at), score: 0.74 UPF2UPF2 regulator of nonsense transcripts homolog (yeast) (203519_s_at), score: 0.65 USP15ubiquitin specific peptidase 15 (210681_s_at), score: 0.76 USP16ubiquitin specific peptidase 16 (218386_x_at), score: 0.67 USP8ubiquitin specific peptidase 8 (202745_at), score: 0.85 VAMP7vesicle-associated membrane protein 7 (202829_s_at), score: 0.85 VGFVGF nerve growth factor inducible (205586_x_at), score: -0.54 VPS13Cvacuolar protein sorting 13 homolog C (S. cerevisiae) (218396_at), score: 0.77 VPS26Avacuolar protein sorting 26 homolog A (S. pombe) (201807_at), score: 0.93 VPS37Bvacuolar protein sorting 37 homolog B (S. cerevisiae) (221704_s_at), score: -0.39 VPS4Bvacuolar protein sorting 4 homolog B (S. cerevisiae) (218171_at), score: 0.82 WBP5WW domain binding protein 5 (217975_at), score: 0.72 WDTC1WD and tetratricopeptide repeats 1 (215497_s_at), score: -0.37 WISP2WNT1 inducible signaling pathway protein 2 (205792_at), score: -0.43 WWC1WW and C2 domain containing 1 (213085_s_at), score: -0.39 WWP1WW domain containing E3 ubiquitin protein ligase 1 (212638_s_at), score: 0.65 YES1v-yes-1 Yamaguchi sarcoma viral oncogene homolog 1 (202932_at), score: 0.83 YPEL5yippee-like 5 (Drosophila) (217783_s_at), score: 0.68 YTHDC1YTH domain containing 1 (212455_at), score: 0.64 YTHDC2YTH domain containing 2 (213077_at), score: 0.76 YTHDF3YTH domain family, member 3 (221749_at), score: 0.82 ZC3H13zinc finger CCCH-type containing 13 (212402_at), score: 0.78 ZC3H14zinc finger CCCH-type containing 14 (213064_at), score: 0.89 ZC4H2zinc finger, C4H2 domain containing (220040_x_at), score: -0.31 ZCCHC6zinc finger, CCHC domain containing 6 (220933_s_at), score: 0.81 ZDHHC17zinc finger, DHHC-type containing 17 (212982_at), score: 0.96 ZDHHC18zinc finger, DHHC-type containing 18 (212860_at), score: -0.35 ZFP36L1zinc finger protein 36, C3H type-like 1 (211965_at), score: -0.35 ZMPSTE24zinc metallopeptidase (STE24 homolog, S. cerevisiae) (202939_at), score: 0.77 ZNF277zinc finger protein 277 (218645_at), score: 0.65 ZNF302zinc finger protein 302 (218490_s_at), score: 0.65 ZNF460zinc finger protein 460 (216279_at), score: -0.31 ZNF589zinc finger protein 589 (210062_s_at), score: -0.32 ZNF665zinc finger protein 665 (220760_x_at), score: 0.65 ZPBPzona pellucida binding protein (207021_at), score: -0.4
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| 10590_WBS.CEL | 2 | 8 | WBS | hgu133plus2 | WBS | WBS 1 |
| ctrl a 08-03.CEL | 1 | 1 | DS-CC | hgu133a | none | DS-CC 1 |
| E-GEOD-4219-raw-cel-1311956321.cel | 9 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| t21a 08-03.CEL | 4 | 1 | DS-CC | hgu133a | Down | DS-CC 4 |
| E-GEOD-4219-raw-cel-1311956614.cel | 18 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-3860-raw-cel-1561690304.cel | 8 | 5 | HGPS | hgu133a | none | GMO8398C |
