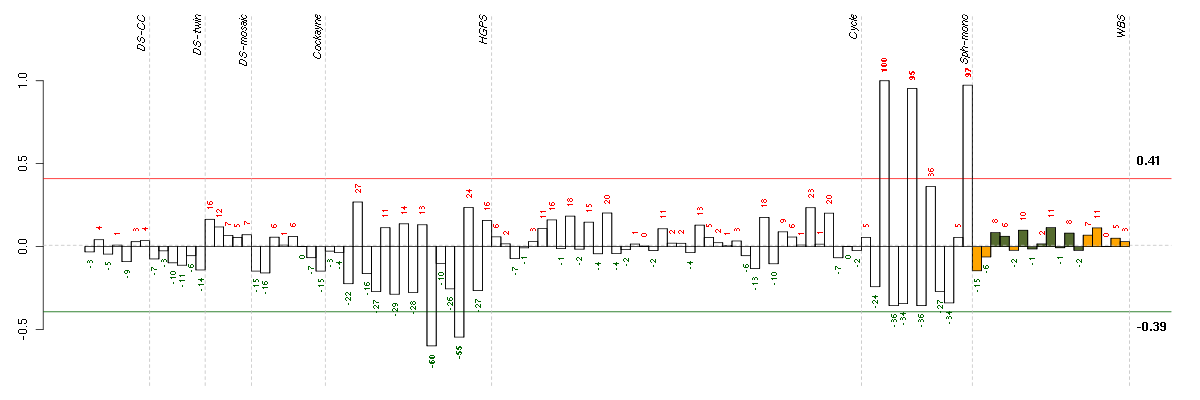

Under-expression is coded with green,
over-expression with red color.


extracellular region
The space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers the host cell environment outside an intracellular parasite.
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
all
This term is the most general term possible

AASSaminoadipate-semialdehyde synthase (214829_at), score: 0.6 ABAT4-aminobutyrate aminotransferase (209459_s_at), score: 0.51 ABCA5ATP-binding cassette, sub-family A (ABC1), member 5 (213353_at), score: 0.54 ACADSacyl-Coenzyme A dehydrogenase, C-2 to C-3 short chain (202366_at), score: -0.46 ACO1aconitase 1, soluble (207071_s_at), score: 0.53 ACTG2actin, gamma 2, smooth muscle, enteric (202274_at), score: 0.51 ADAM19ADAM metallopeptidase domain 19 (meltrin beta) (209765_at), score: 0.5 ADAM9ADAM metallopeptidase domain 9 (meltrin gamma) (202381_at), score: 0.59 ADAMTS1ADAM metallopeptidase with thrombospondin type 1 motif, 1 (222162_s_at), score: 0.64 ADAMTS3ADAM metallopeptidase with thrombospondin type 1 motif, 3 (214913_at), score: 0.81 ADAMTS5ADAM metallopeptidase with thrombospondin type 1 motif, 5 (219935_at), score: -0.46 ADD2adducin 2 (beta) (205268_s_at), score: -0.52 ADIPOR1adiponectin receptor 1 (217748_at), score: -0.52 AEBP1AE binding protein 1 (201792_at), score: 0.69 AGAaspartylglucosaminidase (204332_s_at), score: 0.51 AKAP12A kinase (PRKA) anchor protein 12 (210517_s_at), score: 0.64 AKR1B1aldo-keto reductase family 1, member B1 (aldose reductase) (201272_at), score: -0.54 AKR1C1aldo-keto reductase family 1, member C1 (dihydrodiol dehydrogenase 1; 20-alpha (3-alpha)-hydroxysteroid dehydrogenase) (204151_x_at), score: -0.56 AKR1C2aldo-keto reductase family 1, member C2 (dihydrodiol dehydrogenase 2; bile acid binding protein; 3-alpha hydroxysteroid dehydrogenase, type III) (209699_x_at), score: -0.59 ALCAMactivated leukocyte cell adhesion molecule (201951_at), score: 0.54 ALDH1A1aldehyde dehydrogenase 1 family, member A1 (212224_at), score: 0.89 ANXA3annexin A3 (209369_at), score: 0.62 ANXA8L2annexin A8-like 2 (203074_at), score: -0.45 APAF1apoptotic peptidase activating factor 1 (204859_s_at), score: 0.61 ARF5ADP-ribosylation factor 5 (201526_at), score: -0.47 ARID5BAT rich interactive domain 5B (MRF1-like) (212614_at), score: 0.67 ARL15ADP-ribosylation factor-like 15 (219842_at), score: 0.82 ARMCX1armadillo repeat containing, X-linked 1 (218694_at), score: 0.52 ARMCX3armadillo repeat containing, X-linked 3 (217858_s_at), score: 0.5 ARNTLaryl hydrocarbon receptor nuclear translocator-like (209824_s_at), score: -0.47 ARSJarylsulfatase family, member J (219973_at), score: 0.57 ASAP2ArfGAP with SH3 domain, ankyrin repeat and PH domain 2 (206414_s_at), score: 0.64 ASCC3activating signal cointegrator 1 complex subunit 3 (212815_at), score: 0.5 ASPNasporin (219087_at), score: 0.56 ATF7IP2activating transcription factor 7 interacting protein 2 (219870_at), score: 0.55 ATG2BATG2 autophagy related 2 homolog B (S. cerevisiae) (219164_s_at), score: 0.6 AUP1ancient ubiquitous protein 1 (220525_s_at), score: -0.51 AUTS2autism susceptibility candidate 2 (212599_at), score: -0.61 B3GALT2UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 2 (217452_s_at), score: 0.52 B4GALT5UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 5 (221485_at), score: -0.48 BAZ2Bbromodomain adjacent to zinc finger domain, 2B (203080_s_at), score: 0.51 BCAMbasal cell adhesion molecule (Lutheran blood group) (40093_at), score: 0.54 BCAP31B-cell receptor-associated protein 31 (200837_at), score: -0.59 BEX1brain expressed, X-linked 1 (218332_at), score: 0.51 BIRC2baculoviral IAP repeat-containing 2 (202076_at), score: -0.46 BMP7bone morphogenetic protein 7 (209590_at), score: -0.46 BRIP1BRCA1 interacting protein C-terminal helicase 1 (221703_at), score: 0.54 BSCL2Bernardinelli-Seip congenital lipodystrophy 2 (seipin) (208906_at), score: -0.44 BST1bone marrow stromal cell antigen 1 (205715_at), score: 0.55 BTN3A1butyrophilin, subfamily 3, member A1 (209770_at), score: 0.55 C14orf106chromosome 14 open reading frame 106 (206500_s_at), score: 0.53 C14orf118chromosome 14 open reading frame 118 (219720_s_at), score: 0.51 C14orf169chromosome 14 open reading frame 169 (219526_at), score: 0.65 C15orf29chromosome 15 open reading frame 29 (218791_s_at), score: 0.68 C18orf1chromosome 18 open reading frame 1 (209574_s_at), score: -0.54 C19orf2chromosome 19 open reading frame 2 (214173_x_at), score: -0.56 C20orf111chromosome 20 open reading frame 111 (209020_at), score: -0.48 C20orf39chromosome 20 open reading frame 39 (219310_at), score: -0.44 C21orf2chromosome 21 open reading frame 2 (203996_s_at), score: -0.47 C21orf7chromosome 21 open reading frame 7 (221211_s_at), score: 0.59 C2CD3C2 calcium-dependent domain containing 3 (36552_at), score: 0.57 C2orf37chromosome 2 open reading frame 37 (220172_at), score: 0.62 C4orf31chromosome 4 open reading frame 31 (219747_at), score: -0.45 C6orf211chromosome 6 open reading frame 211 (218195_at), score: 0.67 C7orf23chromosome 7 open reading frame 23 (204215_at), score: -0.49 C7orf28Bchromosome 7 open reading frame 28B (201973_s_at), score: -0.46 C8orf33chromosome 8 open reading frame 33 (218187_s_at), score: -0.59 C9orf125chromosome 9 open reading frame 125 (213386_at), score: 0.5 CABC1chaperone, ABC1 activity of bc1 complex homolog (S. pombe) (218168_s_at), score: -0.45 CALM1calmodulin 1 (phosphorylase kinase, delta) (209563_x_at), score: 0.52 CAND1cullin-associated and neddylation-dissociated 1 (208839_s_at), score: 0.59 CAND2cullin-associated and neddylation-dissociated 2 (putative) (213547_at), score: 0.72 CBFA2T2core-binding factor, runt domain, alpha subunit 2; translocated to, 2 (207625_s_at), score: -0.45 CCL2chemokine (C-C motif) ligand 2 (216598_s_at), score: 0.57 CD24CD24 molecule (209771_x_at), score: -0.65 CD44CD44 molecule (Indian blood group) (210916_s_at), score: -0.77 CDK5RAP2CDK5 regulatory subunit associated protein 2 (220935_s_at), score: -0.51 CELSR3cadherin, EGF LAG seven-pass G-type receptor 3 (flamingo homolog, Drosophila) (40020_at), score: -0.48 CFHcomplement factor H (213800_at), score: 0.58 CGBchorionic gonadotropin, beta polypeptide (205387_s_at), score: -0.55 CH25Hcholesterol 25-hydroxylase (206932_at), score: -0.45 CHCHD2coiled-coil-helix-coiled-coil-helix domain containing 2 (217720_at), score: -0.45 CHMP1Achromatin modifying protein 1A (201933_at), score: -0.48 CHRDL1chordin-like 1 (209763_at), score: 0.65 CLASP2cytoplasmic linker associated protein 2 (212306_at), score: 0.74 CLEC2BC-type lectin domain family 2, member B (209732_at), score: -0.89 COBLL1COBL-like 1 (203642_s_at), score: 0.97 COL4A5collagen, type IV, alpha 5 (213110_s_at), score: 0.6 COL5A2collagen, type V, alpha 2 (221730_at), score: 0.5 COMMD9COMM domain containing 9 (218072_at), score: -0.45 COMPcartilage oligomeric matrix protein (205713_s_at), score: -0.45 CORINcorin, serine peptidase (220356_at), score: 0.6 CORO1Ccoronin, actin binding protein, 1C (221676_s_at), score: 0.51 CORO2Bcoronin, actin binding protein, 2B (209789_at), score: 0.51 COX15COX15 homolog, cytochrome c oxidase assembly protein (yeast) (221550_at), score: 0.5 CPEcarboxypeptidase E (201117_s_at), score: -0.74 CPEB3cytoplasmic polyadenylation element binding protein 3 (205773_at), score: 0.5 CRIM1cysteine rich transmembrane BMP regulator 1 (chordin-like) (202552_s_at), score: 0.6 CSPG4chondroitin sulfate proteoglycan 4 (214297_at), score: 0.58 CTNND1catenin (cadherin-associated protein), delta 1 (208407_s_at), score: 0.51 CTPS2CTP synthase II (219080_s_at), score: 0.57 CTSCcathepsin C (201487_at), score: 0.6 CTSDcathepsin D (200766_at), score: -0.49 CTSScathepsin S (202901_x_at), score: -0.46 CUGBP2CUG triplet repeat, RNA binding protein 2 (202158_s_at), score: 0.55 DAAM1dishevelled associated activator of morphogenesis 1 (216060_s_at), score: 0.51 DAAM2dishevelled associated activator of morphogenesis 2 (212793_at), score: 0.5 DAPK1death-associated protein kinase 1 (203139_at), score: 0.56 DCNdecorin (211896_s_at), score: 0.5 DCUN1D2DCN1, defective in cullin neddylation 1, domain containing 2 (S. cerevisiae) (219116_s_at), score: -0.53 DDAH1dimethylarginine dimethylaminohydrolase 1 (209094_at), score: 0.64 DENND3DENN/MADD domain containing 3 (212975_at), score: 0.62 DENND5BDENN/MADD domain containing 5B (215058_at), score: -0.44 DERL1Der1-like domain family, member 1 (219402_s_at), score: -0.46 DGCR2DiGeorge syndrome critical region gene 2 (214198_s_at), score: -0.45 DGKDdiacylglycerol kinase, delta 130kDa (208072_s_at), score: -0.53 DHDDSdehydrodolichyl diphosphate synthase (218547_at), score: 0.5 DKFZp434H1419hypothetical protein DKFZp434H1419 (214717_at), score: 0.57 DLX2distal-less homeobox 2 (207147_at), score: -0.47 DMDdystrophin (203881_s_at), score: 0.62 DNA2DNA replication helicase 2 homolog (yeast) (213647_at), score: 0.49 DNAJC13DnaJ (Hsp40) homolog, subfamily C, member 13 (212467_at), score: 0.55 DNAJC15DnaJ (Hsp40) homolog, subfamily C, member 15 (218435_at), score: 0.74 DNAJC6DnaJ (Hsp40) homolog, subfamily C, member 6 (204720_s_at), score: 0.61 DRG1developmentally regulated GTP binding protein 1 (202810_at), score: -0.46 DSEdermatan sulfate epimerase (218854_at), score: 0.5 EDEM2ER degradation enhancer, mannosidase alpha-like 2 (218282_at), score: -0.5 EFHD1EF-hand domain family, member D1 (209343_at), score: 0.6 EIF1eukaryotic translation initiation factor 1 (212130_x_at), score: -0.5 EIF2C2eukaryotic translation initiation factor 2C, 2 (213310_at), score: 0.52 EIF3Deukaryotic translation initiation factor 3, subunit D (200005_at), score: -0.48 EML5echinoderm microtubule associated protein like 5 (213063_at), score: 0.55 EMP2epithelial membrane protein 2 (204975_at), score: -0.63 EMX2empty spiracles homeobox 2 (221950_at), score: -0.49 ENOSF1enolase superfamily member 1 (204142_at), score: 0.71 EPHB6EPH receptor B6 (204718_at), score: 0.52 EPHX1epoxide hydrolase 1, microsomal (xenobiotic) (202017_at), score: -0.67 EPS15L1epidermal growth factor receptor pathway substrate 15-like 1 (222113_s_at), score: 0.5 EREGepiregulin (205767_at), score: 0.54 ERGIC3ERGIC and golgi 3 (216032_s_at), score: -0.58 ERP44endoplasmic reticulum protein 44 (208959_s_at), score: -0.48 ESM1endothelial cell-specific molecule 1 (208394_x_at), score: 0.58 ETFBelectron-transfer-flavoprotein, beta polypeptide (202942_at), score: -0.49 ETNK1ethanolamine kinase 1 (219017_at), score: 0.5 EVI2Aecotropic viral integration site 2A (204774_at), score: -0.52 EXOGendo/exonuclease (5'-3'), endonuclease G-like (205521_at), score: 0.57 EXT1exostoses (multiple) 1 (201995_at), score: 0.52 F8coagulation factor VIII, procoagulant component (205756_s_at), score: -0.58 FABP3fatty acid binding protein 3, muscle and heart (mammary-derived growth inhibitor) (214285_at), score: 0.58 FAF1Fas (TNFRSF6) associated factor 1 (218080_x_at), score: 0.53 FAM105Afamily with sequence similarity 105, member A (219694_at), score: -0.46 FAM174Bfamily with sequence similarity 174, member B (51158_at), score: 0.71 FAM46Afamily with sequence similarity 46, member A (221766_s_at), score: -0.51 FAM46Cfamily with sequence similarity 46, member C (220306_at), score: 0.74 FAM65Bfamily with sequence similarity 65, member B (209829_at), score: 0.57 FAR2fatty acyl CoA reductase 2 (220615_s_at), score: 0.61 FEM1Bfem-1 homolog b (C. elegans) (212367_at), score: 0.5 FGF1fibroblast growth factor 1 (acidic) (205117_at), score: 0.62 FGF13fibroblast growth factor 13 (205110_s_at), score: -0.74 FLJ21075hypothetical protein FLJ21075 (221172_at), score: 0.56 FLT1fms-related tyrosine kinase 1 (vascular endothelial growth factor/vascular permeability factor receptor) (222033_s_at), score: 0.81 FOXE1forkhead box E1 (thyroid transcription factor 2) (206912_at), score: 0.68 FOXL2forkhead box L2 (220102_at), score: -0.69 FRYfurry homolog (Drosophila) (204072_s_at), score: 0.58 FYCO1FYVE and coiled-coil domain containing 1 (218204_s_at), score: 0.65 FZD4frizzled homolog 4 (Drosophila) (218665_at), score: 0.73 FZD7frizzled homolog 7 (Drosophila) (203705_s_at), score: 0.55 GAL3ST4galactose-3-O-sulfotransferase 4 (219815_at), score: 0.69 GALNT3UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 3 (GalNAc-T3) (203397_s_at), score: 1 GFRA1GDNF family receptor alpha 1 (205696_s_at), score: 0.49 GIPC2GIPC PDZ domain containing family, member 2 (219970_at), score: 0.53 GPATCH4G patch domain containing 4 (220596_at), score: -0.47 GPR126G protein-coupled receptor 126 (213094_at), score: 0.79 GPR177G protein-coupled receptor 177 (221958_s_at), score: 0.64 GRAMD4GRAM domain containing 4 (212856_at), score: -0.48 GREM1gremlin 1, cysteine knot superfamily, homolog (Xenopus laevis) (218469_at), score: 0.7 GRIK2glutamate receptor, ionotropic, kainate 2 (213845_at), score: 0.53 GULP1GULP, engulfment adaptor PTB domain containing 1 (204237_at), score: 0.57 HEXBhexosaminidase B (beta polypeptide) (201944_at), score: -0.47 HLA-Bmajor histocompatibility complex, class I, B (211911_x_at), score: -0.58 HLA-Cmajor histocompatibility complex, class I, C (211799_x_at), score: -0.51 HLA-Emajor histocompatibility complex, class I, E (200904_at), score: -0.54 HLA-Fmajor histocompatibility complex, class I, F (204806_x_at), score: -0.54 HMGA2high mobility group AT-hook 2 (208025_s_at), score: 0.66 HOMER2homer homolog 2 (Drosophila) (217080_s_at), score: -0.45 HOXB5homeobox B5 (205601_s_at), score: -0.5 HOXD4homeobox D4 (205522_at), score: -0.44 HSD17B12hydroxysteroid (17-beta) dehydrogenase 12 (217869_at), score: 0.53 IDH3Bisocitrate dehydrogenase 3 (NAD+) beta (210014_x_at), score: -0.44 IGF2insulin-like growth factor 2 (somatomedin A) (202409_at), score: 0.59 IL23Ainterleukin 23, alpha subunit p19 (211796_s_at), score: 0.79 IMAASLC7A5 pseudogene (208118_x_at), score: -0.62 INAinternexin neuronal intermediate filament protein, alpha (204465_s_at), score: 0.79 INHBEinhibin, beta E (210587_at), score: 0.56 INPP5Ainositol polyphosphate-5-phosphatase, 40kDa (203006_at), score: 0.54 IPWimprinted in Prader-Willi syndrome (non-protein coding) (221974_at), score: 0.61 IRX5iroquois homeobox 5 (210239_at), score: -0.51 ISL1ISL LIM homeobox 1 (206104_at), score: 0.73 ITGAVintegrin, alpha V (vitronectin receptor, alpha polypeptide, antigen CD51) (202351_at), score: 0.58 KAL1Kallmann syndrome 1 sequence (205206_at), score: 0.59 KBTBD11kelch repeat and BTB (POZ) domain containing 11 (204301_at), score: -0.45 KCNJ2potassium inwardly-rectifying channel, subfamily J, member 2 (206765_at), score: 0.76 KCNK2potassium channel, subfamily K, member 2 (210261_at), score: 0.55 KCNN2potassium intermediate/small conductance calcium-activated channel, subfamily N, member 2 (220116_at), score: 0.62 KCTD2potassium channel tetramerisation domain containing 2 (34858_at), score: -0.47 KHDRBS3KH domain containing, RNA binding, signal transduction associated 3 (209781_s_at), score: 0.52 KIAA0802KIAA0802 (213358_at), score: 0.6 KIAA1199KIAA1199 (212942_s_at), score: 0.5 KIF3Bkinesin family member 3B (203943_at), score: -0.46 KLHL3kelch-like 3 (Drosophila) (221221_s_at), score: 0.59 KYNUkynureninase (L-kynurenine hydrolase) (217388_s_at), score: -0.67 L1CAML1 cell adhesion molecule (204584_at), score: 0.72 LAPTM5lysosomal multispanning membrane protein 5 (201721_s_at), score: -0.64 LEF1lymphoid enhancer-binding factor 1 (221558_s_at), score: -0.44 LHFPlipoma HMGIC fusion partner (218656_s_at), score: 0.51 LIN7Alin-7 homolog A (C. elegans) (206440_at), score: 0.57 LMO7LIM domain 7 (202674_s_at), score: 0.73 LOC220594TL132 protein (213510_x_at), score: 0.51 LOC440354PI-3-kinase-related kinase SMG-1 pseudogene (210396_s_at), score: -0.48 LPPR4plasticity related gene 1 (213496_at), score: 0.85 LRP4low density lipoprotein receptor-related protein 4 (212850_s_at), score: -0.5 LRRC1leucine rich repeat containing 1 (218816_at), score: 0.64 LRRC16Aleucine rich repeat containing 16A (219573_at), score: 0.8 LRRC17leucine rich repeat containing 17 (205381_at), score: 0.57 LRRC2leucine rich repeat containing 2 (219949_at), score: 0.56 LRRN3leucine rich repeat neuronal 3 (209841_s_at), score: 0.59 LUMlumican (201744_s_at), score: 0.58 MAFBv-maf musculoaponeurotic fibrosarcoma oncogene homolog B (avian) (218559_s_at), score: -0.54 MAGI2membrane associated guanylate kinase, WW and PDZ domain containing 2 (209737_at), score: 0.54 MAMLD1mastermind-like domain containing 1 (205088_at), score: 0.57 MAN1A2mannosidase, alpha, class 1A, member 2 (217921_at), score: 0.58 MANSC1MANSC domain containing 1 (220945_x_at), score: 0.96 MAP9microtubule-associated protein 9 (220145_at), score: 0.52 MAPK13mitogen-activated protein kinase 13 (210059_s_at), score: 0.63 MARCH3membrane-associated ring finger (C3HC4) 3 (213256_at), score: 0.56 MBNL1muscleblind-like (Drosophila) (201151_s_at), score: 0.52 ME1malic enzyme 1, NADP(+)-dependent, cytosolic (204058_at), score: 0.7 ME3malic enzyme 3, NADP(+)-dependent, mitochondrial (204663_at), score: 0.5 MECP2methyl CpG binding protein 2 (Rett syndrome) (202616_s_at), score: -0.44 MED21mediator complex subunit 21 (209362_at), score: 0.53 MED23mediator complex subunit 23 (218846_at), score: 0.53 MKRN2makorin ring finger protein 2 (218071_s_at), score: 0.61 MLF1myeloid leukemia factor 1 (204784_s_at), score: -0.59 MLLT3myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 3 (204918_s_at), score: 0.85 MMP2matrix metallopeptidase 2 (gelatinase A, 72kDa gelatinase, 72kDa type IV collagenase) (201069_at), score: -0.5 MRPL52mitochondrial ribosomal protein L52 (221997_s_at), score: 0.57 MRPS18Cmitochondrial ribosomal protein S18C (220103_s_at), score: 0.57 MYO1Bmyosin IB (212364_at), score: 0.67 NACC2NACC family member 2, BEN and BTB (POZ) domain containing (212993_at), score: 0.54 NAV3neuron navigator 3 (204823_at), score: 0.62 NBEAneurobeachin (221207_s_at), score: 0.69 NBL1neuroblastoma, suppression of tumorigenicity 1 (37005_at), score: -0.53 NCAM1neural cell adhesion molecule 1 (212843_at), score: -0.49 NDPNorrie disease (pseudoglioma) (206022_at), score: -0.53 NEDD9neural precursor cell expressed, developmentally down-regulated 9 (202149_at), score: 0.65 NESnestin (218678_at), score: 0.77 NID2nidogen 2 (osteonidogen) (204114_at), score: 0.64 NLGN1neuroligin 1 (205893_at), score: 0.63 NLKnemo-like kinase (218318_s_at), score: 0.54 NOVnephroblastoma overexpressed gene (214321_at), score: 0.63 NPAS2neuronal PAS domain protein 2 (39549_at), score: 0.55 NPC1Niemann-Pick disease, type C1 (202679_at), score: -0.54 NPLOC4nuclear protein localization 4 homolog (S. cerevisiae) (217796_s_at), score: -0.47 NR2F2nuclear receptor subfamily 2, group F, member 2 (215073_s_at), score: 0.7 NRASneuroblastoma RAS viral (v-ras) oncogene homolog (202647_s_at), score: 0.51 NRN1neuritin 1 (218625_at), score: -0.7 NUAK2NUAK family, SNF1-like kinase, 2 (220987_s_at), score: 0.65 OBFC1oligonucleotide/oligosaccharide-binding fold containing 1 (219100_at), score: -0.46 OPTNoptineurin (202073_at), score: 0.52 OSBPL10oxysterol binding protein-like 10 (219073_s_at), score: 0.66 PARGpoly (ADP-ribose) glycohydrolase (205060_at), score: 0.57 PBXIP1pre-B-cell leukemia homeobox interacting protein 1 (214176_s_at), score: 0.61 PCGF1polycomb group ring finger 1 (210023_s_at), score: -0.48 PEX5peroxisomal biogenesis factor 5 (203244_at), score: -0.57 PFDN2prefoldin subunit 2 (218336_at), score: -0.49 PFTK1PFTAIRE protein kinase 1 (204604_at), score: 0.51 PGAP1post-GPI attachment to proteins 1 (213469_at), score: 0.53 PHLDA2pleckstrin homology-like domain, family A, member 2 (209803_s_at), score: 0.55 PLAGL1pleiomorphic adenoma gene-like 1 (209318_x_at), score: 0.58 PLCB1phospholipase C, beta 1 (phosphoinositide-specific) (213222_at), score: 0.76 PLCXD1phosphatidylinositol-specific phospholipase C, X domain containing 1 (218951_s_at), score: 0.5 PLP1proteolipid protein 1 (210198_s_at), score: 0.65 PLS3plastin 3 (T isoform) (201215_at), score: 0.56 PMS2L11postmeiotic segregation increased 2-like 11 pseudogene (210707_x_at), score: -0.46 POLE2polymerase (DNA directed), epsilon 2 (p59 subunit) (205909_at), score: 0.53 POLR2Fpolymerase (RNA) II (DNA directed) polypeptide F (209511_at), score: -0.44 POLR2J2polymerase (RNA) II (DNA directed) polypeptide J2 (216242_x_at), score: -0.56 POM121CPOM121 membrane glycoprotein C (213360_s_at), score: -0.52 POSTNperiostin, osteoblast specific factor (210809_s_at), score: 0.5 PPIBpeptidylprolyl isomerase B (cyclophilin B) (200968_s_at), score: -0.52 PPP3CBprotein phosphatase 3 (formerly 2B), catalytic subunit, beta isoform (209817_at), score: 0.51 PPP3R1protein phosphatase 3 (formerly 2B), regulatory subunit B, alpha isoform (204507_s_at), score: 0.49 PRCCpapillary renal cell carcinoma (translocation-associated) (208938_at), score: -0.47 PRPF38BPRP38 pre-mRNA processing factor 38 (yeast) domain containing B (218040_at), score: 0.51 PRR7proline rich 7 (synaptic) (219742_at), score: -0.46 PRSS1protease, serine, 1 (trypsin 1) (216470_x_at), score: 0.53 PSMB3proteasome (prosome, macropain) subunit, beta type, 3 (201400_at), score: -0.48 PSMD5proteasome (prosome, macropain) 26S subunit, non-ATPase, 5 (203447_at), score: 0.55 PSTPIP2proline-serine-threonine phosphatase interacting protein 2 (219938_s_at), score: 0.67 PTPRDprotein tyrosine phosphatase, receptor type, D (214043_at), score: 0.61 PTX3pentraxin-related gene, rapidly induced by IL-1 beta (206157_at), score: 0.52 PYROXD1pyridine nucleotide-disulphide oxidoreductase domain 1 (213878_at), score: 0.53 RAB11BRAB11B, member RAS oncogene family (34478_at), score: -0.48 RAB31RAB31, member RAS oncogene family (217763_s_at), score: -0.57 RAB33BRAB33B, member RAS oncogene family (221014_s_at), score: 0.53 RAD23BRAD23 homolog B (S. cerevisiae) (201222_s_at), score: 0.5 RAI14retinoic acid induced 14 (202052_s_at), score: 0.6 RB1retinoblastoma 1 (203132_at), score: 0.51 RBM47RNA binding motif protein 47 (218035_s_at), score: -0.47 RCAN2regulator of calcineurin 2 (203498_at), score: 0.54 RECKreversion-inducing-cysteine-rich protein with kazal motifs (205407_at), score: 0.52 RGS4regulator of G-protein signaling 4 (204337_at), score: 0.78 RGS7regulator of G-protein signaling 7 (206290_s_at), score: 0.51 RHOBras homolog gene family, member B (212099_at), score: 0.59 RIF1RAP1 interacting factor homolog (yeast) (214700_x_at), score: 0.53 RIPK4receptor-interacting serine-threonine kinase 4 (221215_s_at), score: -0.62 ROD1ROD1 regulator of differentiation 1 (S. pombe) (214697_s_at), score: 0.58 RPL30ribosomal protein L30 (200062_s_at), score: -0.46 RPLP0P6ribosomal protein, large, P0 pseudogene 6 (214167_s_at), score: -0.46 RPRD1Aregulation of nuclear pre-mRNA domain containing 1A (218209_s_at), score: 0.58 RPS4Xribosomal protein S4, X-linked (200933_x_at), score: -0.48 RPS6KA5ribosomal protein S6 kinase, 90kDa, polypeptide 5 (204633_s_at), score: -0.47 SALL1sal-like 1 (Drosophila) (206893_at), score: -0.53 SALL2sal-like 2 (Drosophila) (213283_s_at), score: 0.55 SAP18Sin3A-associated protein, 18kDa (208740_at), score: -0.51 SAP30BPSAP30 binding protein (217965_s_at), score: -0.48 SCG5secretogranin V (7B2 protein) (203889_at), score: -0.46 SCN3Asodium channel, voltage-gated, type III, alpha subunit (210432_s_at), score: 0.71 SCN9Asodium channel, voltage-gated, type IX, alpha subunit (206950_at), score: 0.8 SCRIBscribbled homolog (Drosophila) (212556_at), score: 0.63 SDCCAG1serologically defined colon cancer antigen 1 (218649_x_at), score: 0.61 SEC22BSEC22 vesicle trafficking protein homolog B (S. cerevisiae) (214257_s_at), score: 0.53 SERPINB7serpin peptidase inhibitor, clade B (ovalbumin), member 7 (206421_s_at), score: 0.51 SERPINB9serpin peptidase inhibitor, clade B (ovalbumin), member 9 (209723_at), score: 0.84 SFTPBsurfactant protein B (214354_x_at), score: -0.45 SGK1serum/glucocorticoid regulated kinase 1 (201739_at), score: 0.68 SHROOM2shroom family member 2 (204967_at), score: 0.53 SIRT3sirtuin (silent mating type information regulation 2 homolog) 3 (S. cerevisiae) (221562_s_at), score: -0.56 SKP2S-phase kinase-associated protein 2 (p45) (203625_x_at), score: 0.53 SLC20A2solute carrier family 20 (phosphate transporter), member 2 (202744_at), score: 0.65 SLC24A1solute carrier family 24 (sodium/potassium/calcium exchanger), member 1 (206081_at), score: 0.61 SLC35F5solute carrier family 35, member F5 (220123_at), score: 0.63 SLC38A4solute carrier family 38, member 4 (220786_s_at), score: 0.76 SLC3A1solute carrier family 3 (cystine, dibasic and neutral amino acid transporters, activator of cystine, dibasic and neutral amino acid transport), member 1 (205799_s_at), score: 0.75 SLC48A1solute carrier family 48 (heme transporter), member 1 (218417_s_at), score: 0.54 SMA5glucuronidase, beta pseudogene (215043_s_at), score: -0.58 SMAD4SMAD family member 4 (202527_s_at), score: 0.5 SMARCE1SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily e, member 1 (211989_at), score: -0.45 SMURF2SMAD specific E3 ubiquitin protein ligase 2 (205596_s_at), score: 0.66 SNF8SNF8, ESCRT-II complex subunit, homolog (S. cerevisiae) (218391_at), score: -0.5 SNRPD1small nuclear ribonucleoprotein D1 polypeptide 16kDa (202691_at), score: 0.53 SPCS3signal peptidase complex subunit 3 homolog (S. cerevisiae) (218817_at), score: 0.5 SPHARS-phase response (cyclin-related) (206272_at), score: 0.56 ST3GAL5ST3 beta-galactoside alpha-2,3-sialyltransferase 5 (203217_s_at), score: 0.51 ST6GAL1ST6 beta-galactosamide alpha-2,6-sialyltranferase 1 (201998_at), score: 0.57 STAM2signal transducing adaptor molecule (SH3 domain and ITAM motif) 2 (209649_at), score: 0.59 STARD3StAR-related lipid transfer (START) domain containing 3 (202991_at), score: -0.57 STAU2staufen, RNA binding protein, homolog 2 (Drosophila) (204226_at), score: 0.61 STK16serine/threonine kinase 16 (209622_at), score: -0.45 STK39serine threonine kinase 39 (STE20/SPS1 homolog, yeast) (202786_at), score: 0.64 STOMstomatin (201060_x_at), score: 0.53 STRN3striatin, calmodulin binding protein 3 (204496_at), score: 0.5 STSsteroid sulfatase (microsomal), isozyme S (203767_s_at), score: 0.54 STX2syntaxin 2 (213434_at), score: 0.55 STX7syntaxin 7 (212631_at), score: 0.66 SULF1sulfatase 1 (212353_at), score: 0.52 SVILsupervillin (202565_s_at), score: 0.53 SYNGR1synaptogyrin 1 (210613_s_at), score: 0.61 SYNMsynemin, intermediate filament protein (212730_at), score: 0.73 SYT11synaptotagmin XI (209198_s_at), score: 0.68 TBC1D2BTBC1 domain family, member 2B (212796_s_at), score: 0.67 TBC1D3TBC1 domain family, member 3 (209403_at), score: -0.45 TBC1D9TBC1 domain family, member 9 (with GRAM domain) (212956_at), score: 0.5 TBXA2Rthromboxane A2 receptor (336_at), score: 0.91 TEAD3TEA domain family member 3 (209454_s_at), score: 0.52 TEStestis derived transcript (3 LIM domains) (202720_at), score: 0.67 TGFBItransforming growth factor, beta-induced, 68kDa (201506_at), score: -0.57 THAP1THAP domain containing, apoptosis associated protein 1 (219292_at), score: 0.53 THAP10THAP domain containing 10 (219596_at), score: 0.51 THAP4THAP domain containing 4 (220417_s_at), score: -0.49 THBS2thrombospondin 2 (203083_at), score: 0.69 THSD1thrombospondin, type I, domain containing 1 (219477_s_at), score: -0.44 TMBIM1transmembrane BAX inhibitor motif containing 1 (217730_at), score: -0.58 TMEM111transmembrane protein 111 (217882_at), score: -0.66 TMEM135transmembrane protein 135 (222209_s_at), score: 0.52 TMEM158transmembrane protein 158 (213338_at), score: -0.47 TMEM168transmembrane protein 168 (218962_s_at), score: 0.64 TMEM2transmembrane protein 2 (218113_at), score: 0.69 TMEM30Btransmembrane protein 30B (213285_at), score: 0.51 TMEM35transmembrane protein 35 (219685_at), score: 0.54 TMEM45Atransmembrane protein 45A (219410_at), score: 0.57 TMEM70transmembrane protein 70 (219449_s_at), score: -0.45 TNFAIP8tumor necrosis factor, alpha-induced protein 8 (210260_s_at), score: 0.57 TNFSF4tumor necrosis factor (ligand) superfamily, member 4 (207426_s_at), score: 0.5 TNS1tensin 1 (221748_s_at), score: 0.69 TOXthymocyte selection-associated high mobility group box (204529_s_at), score: 0.76 TRAF5TNF receptor-associated factor 5 (204352_at), score: 0.54 TRIM21tripartite motif-containing 21 (204804_at), score: 0.52 TRIM32tripartite motif-containing 32 (203846_at), score: 0.57 TRIM5tripartite motif-containing 5 (210705_s_at), score: 0.51 TRNAG6transfer RNA glycine 6 (anticodon GCC) (217542_at), score: 0.61 TRPC3transient receptor potential cation channel, subfamily C, member 3 (210814_at), score: -0.45 TSPAN14tetraspanin 14 (221002_s_at), score: 0.5 TSPAN4tetraspanin 4 (209264_s_at), score: -0.44 TSPAN5tetraspanin 5 (209890_at), score: 0.51 TSPYL1TSPY-like 1 (221493_at), score: 0.56 TTC33tetratricopeptide repeat domain 33 (219421_at), score: 0.65 TUFT1tuftelin 1 (205807_s_at), score: 0.56 UBE2Bubiquitin-conjugating enzyme E2B (RAD6 homolog) (211763_s_at), score: 0.49 UBR2ubiquitin protein ligase E3 component n-recognin 2 (212760_at), score: 0.51 UGDHUDP-glucose dehydrogenase (203343_at), score: 0.7 ULK2unc-51-like kinase 2 (C. elegans) (204062_s_at), score: 0.56 UTP18UTP18, small subunit (SSU) processome component, homolog (yeast) (203721_s_at), score: -0.46 VGLL3vestigial like 3 (Drosophila) (220327_at), score: 0.65 VPS13Avacuolar protein sorting 13 homolog A (S. cerevisiae) (214785_at), score: 0.62 WDR44WD repeat domain 44 (219297_at), score: 0.5 WFDC8WAP four-disulfide core domain 8 (215276_at), score: -0.47 YIPF4Yip1 domain family, member 4 (209551_at), score: 0.52 YTHDC2YTH domain containing 2 (213077_at), score: 0.6 ZCCHC14zinc finger, CCHC domain containing 14 (212655_at), score: -0.51 ZMIZ1zinc finger, MIZ-type containing 1 (212124_at), score: -0.55 ZNF140zinc finger protein 140 (204523_at), score: 0.5 ZNF215zinc finger protein 215 (220214_at), score: 0.64 ZNF365zinc finger protein 365 (206448_at), score: 0.5 ZNF518Azinc finger protein 518A (204291_at), score: 0.6
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-GEOD-3860-raw-cel-1561690360.cel | 12 | 5 | HGPS | hgu133a | none | GM0316B |
| E-GEOD-3860-raw-cel-1561690416.cel | 15 | 5 | HGPS | hgu133a | none | GM0316B |
| E-GEOD-4219-raw-cel-1311956358.cel | 10 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956824.cel | 24 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956178.cel | 6 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |