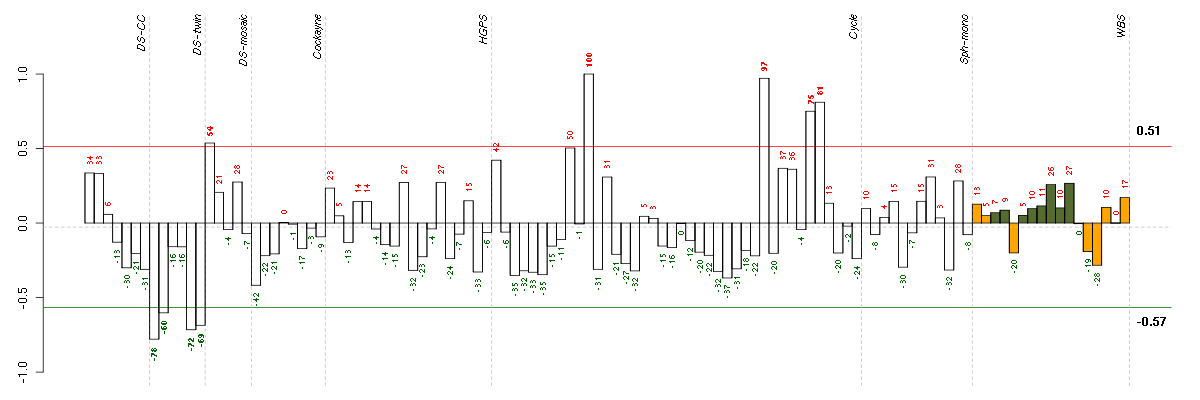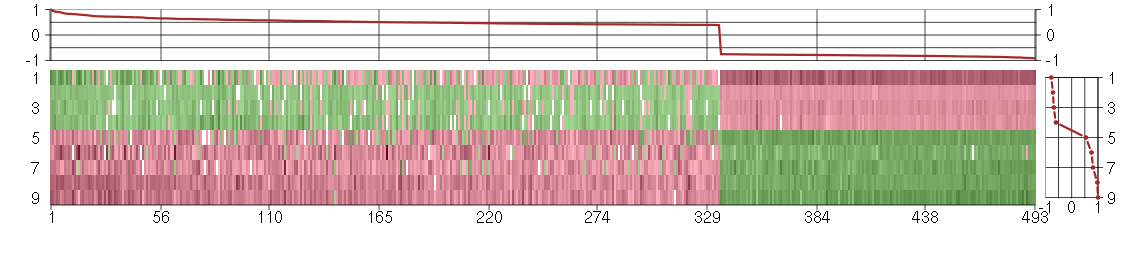

Under-expression is coded with green,
over-expression with red color.

DNA replication
The process whereby new strands of DNA are synthesized. The template for replication can either be an existing DNA molecule or RNA.
chromosome segregation
The process by which genetic material, in the form of chromosomes, is organized and then physically separated and apportioned to two or more sets.
chromosome organization
A process that is carried out at the cellular level that results in the formation, arrangement of constituent parts, or disassembly of chromosomes, structures composed of a very long molecule of DNA and associated proteins that carries hereditary information.
mitotic sister chromatid segregation
The cell cycle process whereby replicated homologous chromosomes are organized and then physically separated and apportioned to two sets during the mitotic cell cycle. Each replicated chromosome, composed of two sister chromatids, aligns at the cell equator, paired with its homologous partner. One homolog of each morphologic type goes into each of the resulting chromosome sets.
cell cycle checkpoint
The cell cycle regulatory process by which progression through the cycle can be halted until conditions are suitable for the cell to proceed to the next stage.
M phase of mitotic cell cycle
A cell cycle process comprising the steps by which a cell progresses through M phase, the part of the mitotic cell cycle during which mitosis takes place.
microtubule cytoskeleton organization
A process that is carried out at the cellular level which results in the formation, arrangement of constituent parts, or disassembly of cytoskeletal structures comprising microtubules and their associated proteins.
mitotic cell cycle
Progression through the phases of the mitotic cell cycle, the most common eukaryotic cell cycle, which canonically comprises four successive phases called G1, S, G2, and M and includes replication of the genome and the subsequent segregation of chromosomes into daughter cells. In some variant cell cycles nuclear replication or nuclear division may not be followed by cell division, or G1 and G2 phases may be absent.
M phase
A cell cycle process comprising the steps by which a cell progresses through M phase, the part of the cell cycle comprising nuclear division.
nuclear division
A process by which a cell nucleus is divided into two nuclei, with DNA and other nuclear contents distributed between the daughter nuclei.
double-strand break repair via homologous recombination
The error-free repair of a double-strand break in DNA in which the broken DNA molecule is repaired using homologous sequences. A strand in the broken DNA searches for a homologous region in an intact chromosome to serve as the template for DNA synthesis. The restoration of two intact DNA molecules results in the exchange, reciprocal or nonreciprocal, of genetic material between the intact DNA molecule and the broken DNA molecule.
recombinational repair
The repair of damaged DNA that involves the exchange, reciprocal or nonreciprocal, of genetic material between the broken DNA molecule and a homologous region of DNA.
DNA synthesis during DNA repair
Synthesis of DNA that proceeds from the broken 3' single-strand DNA end uses the homologous intact duplex as the template.
sister chromatid segregation
The process by which sister chromatids are organized and then physically separated and apportioned to two or more sets.
skeletal system development
The process whose specific outcome is the progression of the skeleton over time, from its formation to the mature structure. The skeleton is the bony framework of the body in vertebrates (endoskeleton) or the hard outer envelope of insects (exoskeleton or dermoskeleton).
ossification
The formation of bone or of a bony substance, or the conversion of fibrous tissue or of cartilage into bone or a bony substance.
angiogenesis
Blood vessel formation when new vessels emerge from the proliferation of pre-existing blood vessels.
metabolic process
The chemical reactions and pathways, including anabolism and catabolism, by which living organisms transform chemical substances. Metabolic processes typically transform small molecules, but also include macromolecular processes such as DNA repair and replication, and protein synthesis and degradation.
blood vessel development
The process whose specific outcome is the progression of the blood vessel over time, from its formation to the mature structure. The blood vessel is the vasculature carrying blood.
osteoblast differentiation
The process whereby a relatively unspecialized cell acquires the specialized features of an osteoblast, the mesodermal cell that gives rise to bone.
cell activation
A change in the morphology or behavior of a cell resulting from exposure to an activating factor such as a cellular or soluble ligand.
regulation of protein amino acid phosphorylation
Any process that modulates the frequency, rate or extent of addition of phosphate groups into an amino acid in a protein.
vasculature development
The process whose specific outcome is the progression of the vasculature over time, from its formation to the mature structure.
immune system process
Any process involved in the development or functioning of the immune system, an organismal system for calibrated responses to potential internal or invasive threats.
generation of a signal involved in cell-cell signaling
The cellular process by which a physical entity or change in state, a signal, is created that originates in one cell and is used to transfer information to another cell. This process begins with the initial formation of the signal and ends with the mature form and placement of the signal.
regulation of cell cycle
Any process that modulates the rate or extent of progression through the cell cycle.
cytoskeleton organization
A process that is carried out at the cellular level which results in the formation, arrangement of constituent parts, or disassembly of cytoskeletal structures.
secretion
The controlled release of a substance by a cell, a group of cells, or a tissue.
signal transduction
The cascade of processes by which a signal interacts with a receptor, causing a change in the level or activity of a second messenger or other downstream target, and ultimately effecting a change in the functioning of the cell.
cellular alcohol metabolic process
The chemical reactions and pathways involving alcohols, any of a class of compounds containing one or more hydroxyl groups attached to a saturated carbon atom, as carried out by individual cells.
nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
The chemical reactions and pathways involving nucleobases, nucleosides, nucleotides and nucleic acids.
DNA metabolic process
The chemical reactions and pathways involving DNA, deoxyribonucleic acid, one of the two main types of nucleic acid, consisting of a long, unbranched macromolecule formed from one, or more commonly, two, strands of linked deoxyribonucleotides.
DNA-dependent DNA replication
The process whereby new strands of DNA are synthesized, using parental DNA as a template for the DNA-dependent DNA polymerases that synthesize the new strands.
DNA replication initiation
The process by which DNA replication is started; this involves the separation of a stretch of the DNA double helix, the recruitment of DNA polymerases and the initiation of polymerase action.
regulation of DNA replication
Any process that modulates the frequency, rate or extent of DNA replication.
DNA repair
The process of restoring DNA after damage. Genomes are subject to damage by chemical and physical agents in the environment (e.g. UV and ionizing radiations, chemical mutagens, fungal and bacterial toxins, etc.) and by free radicals or alkylating agents endogenously generated in metabolism. DNA is also damaged because of errors during its replication. A variety of different DNA repair pathways have been reported that include direct reversal, base excision repair, nucleotide excision repair, photoreactivation, bypass, double-strand break repair pathway, and mismatch repair pathway.
nucleotide-excision repair
In nucleotide excision repair a small region of the strand surrounding the damage is removed from the DNA helix as an oligonucleotide. The small gap left in the DNA helix is filled in by the sequential action of DNA polymerase and DNA ligase. Nucleotide excision repair recognizes a wide range of substrates, including damage caused by UV irradiation (pyrimidine dimers and 6-4 photoproducts) and chemicals (intrastrand cross-links and bulky adducts).
nucleotide-excision repair, DNA gap filling
Repair of the gap in the DNA helix by DNA polymerase and DNA ligase after the portion of the strand containing the lesion has been removed by pyrimidine-dimer repair enzymes.
double-strand break repair
The repair of double-strand breaks in DNA via homologous and nonhomologous mechanisms to reform a continuous DNA helix.
DNA recombination
Any process by which a new genotype is formed by reassortment of genes resulting in gene combinations different from those that were present in the parents. In eukaryotes genetic recombination can occur by chromosome assortment, intrachromosomal recombination, or nonreciprocal interchromosomal recombination. Intrachromosomal recombination occurs by crossing over. In bacteria it may occur by genetic transformation, conjugation, transduction, or F-duction.
DNA packaging
Any process by which DNA and associated proteins are formed into a compact, orderly structure.
protein modification process
The covalent alteration of one or more amino acids occurring in proteins, peptides and nascent polypeptides (co-translational, post-translational modifications). Includes the modification of charged tRNAs that are destined to occur in a protein (pre-translation modification).
protein amino acid phosphorylation
The process of introducing a phosphate group on to a protein.
lipid metabolic process
The chemical reactions and pathways involving lipids, compounds soluble in an organic solvent but not, or sparingly, in an aqueous solvent. Includes fatty acids; neutral fats, other fatty-acid esters, and soaps; long-chain (fatty) alcohols and waxes; sphingoids and other long-chain bases; glycolipids, phospholipids and sphingolipids; and carotenes, polyprenols, sterols, terpenes and other isoprenoids.
phosphorus metabolic process
The chemical reactions and pathways involving the nonmetallic element phosphorus or compounds that contain phosphorus, usually in the form of a phosphate group (PO4).
phosphate metabolic process
The chemical reactions and pathways involving the phosphate group, the anion or salt of any phosphoric acid.
transport
The directed movement of substances (such as macromolecules, small molecules, ions) into, out of, within or between cells, or within a multicellular organism.
cell motion
Any process involved in the controlled movement of a cell.
response to stress
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a disturbance in organismal or cellular homeostasis, usually, but not necessarily, exogenous (e.g. temperature, humidity, ionizing radiation).
response to DNA damage stimulus
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus indicating damage to its DNA from environmental insults or errors during metabolism.
organelle organization
A process that is carried out at the cellular level which results in the formation, arrangement of constituent parts, or disassembly of an organelle within a cell. An organelle is an organized structure of distinctive morphology and function. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton. Excludes the plasma membrane.
microtubule-based process
Any cellular process that depends upon or alters the microtubule cytoskeleton, that part of the cytoskeleton comprising microtubules and their associated proteins.
cell cycle
The progression of biochemical and morphological phases and events that occur in a cell during successive cell replication or nuclear replication events. Canonically, the cell cycle comprises the replication and segregation of genetic material followed by the division of the cell, but in endocycles or syncytial cells nuclear replication or nuclear division may not be followed by cell division.
mitosis
A cell cycle process comprising the steps by which the nucleus of a eukaryotic cell divides; the process involves condensation of chromosomal DNA into a highly compacted form. Canonically, mitosis produces two daughter nuclei whose chromosome complement is identical to that of the mother cell.
mitotic chromosome condensation
The cell cycle process whereby chromatin structure is compacted prior to mitosis in eukaryotic cells.
regulation of mitosis
Any process that modulates the frequency, rate or extent of mitosis.
cell communication
Any process that mediates interactions between a cell and its surroundings. Encompasses interactions such as signaling or attachment between one cell and another cell, between a cell and an extracellular matrix, or between a cell and any other aspect of its environment.
intracellular signaling cascade
A series of reactions within the cell that occur as a result of a single trigger reaction or compound.
cell-cell signaling
Any process that mediates the transfer of information from one cell to another.
multicellular organismal development
The biological process whose specific outcome is the progression of a multicellular organism over time from an initial condition (e.g. a zygote or a young adult) to a later condition (e.g. a multicellular animal or an aged adult).
anatomical structure morphogenesis
The process by which anatomical structures are generated and organized. Morphogenesis pertains to the creation of form.
regulation of mitotic cell cycle
Any process that modulates the rate or extent of progress through the mitotic cell cycle.
organ morphogenesis
Morphogenesis of an organ. An organ is defined as a tissue or set of tissues that work together to perform a specific function or functions. Morphogenesis is the process by which anatomical structures are generated and organized. Organs are commonly observed as visibly distinct structures, but may also exist as loosely associated clusters of cells that work together to perform a specific function or functions.
biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
steroid metabolic process
The chemical reactions and pathways involving steroids, compounds with a 1,2,cyclopentanoperhydrophenanthrene nucleus.
cell proliferation
The multiplication or reproduction of cells, resulting in the expansion of a cell population.
positive regulation of cell proliferation
Any process that activates or increases the rate or extent of cell proliferation.
biosynthetic process
The chemical reactions and pathways resulting in the formation of substances; typically the energy-requiring part of metabolism in which simpler substances are transformed into more complex ones.
macromolecule biosynthetic process
The chemical reactions and pathways resulting in the formation of a macromolecule, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass.
response to external stimulus
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of an external stimulus.
response to wounding
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus indicating damage to the organism.
regulation of biosynthetic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways resulting in the formation of substances.
positive regulation of metabolic process
Any process that activates or increases the frequency, rate or extent of the chemical reactions and pathways within a cell or an organism.
hormone transport
The directed movement of hormones into, out of, within or between cells.
cellular process
Any process that is carried out at the cellular level, but not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
regulation of macromolecule biosynthetic process
Any process that modulates the rate, frequency or extent of the chemical reactions and pathways resulting in the formation of a macromolecule, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass.
positive regulation of phosphorus metabolic process
Any process that increases the frequency, rate or extent of the chemical reactions and pathways involving phosphorus or compounds containing phosphorus.
regulation of cell cycle process
Any process that modulates a cellular process that is involved in the progression of biochemical and morphological phases and events that occur in a cell during successive cell replication or nuclear replication events.
positive regulation of macromolecule metabolic process
Any process that increases the frequency, rate or extent of the chemical reactions and pathways involving macromolecules, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass.
regulation of cell communication
Any process that modulates the frequency, rate or extent of cell communication. Cell communication is the process that mediates interactions between a cell and its surroundings. Encompasses interactions such as signaling or attachment between one cell and another cell, between a cell and an extracellular matrix, or between a cell and any other aspect of its environment.
regulation of hormone levels
Any process that modulates the levels of hormone within an organism or a tissue. A hormone is any substance formed in very small amounts in one specialized organ or group of cells and carried (sometimes in the bloodstream) to another organ or group of cells in the same organism, upon which it has a specific regulatory action.
cellular component organization
A process that is carried out at the cellular level which results in the formation, arrangement of constituent parts, or disassembly of a cellular component.
sterol metabolic process
The chemical reactions and pathways involving sterols, steroids with one or more hydroxyl groups and a hydrocarbon side-chain in the molecule.
phosphorylation
The process of introducing a phosphate group into a molecule, usually with the formation of a phosphoric ester, a phosphoric anhydride or a phosphoric amide.
cell migration
The orderly movement of cells from one site to another, often during the development of a multicellular organism or multicellular structure.
regulation of nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways involving nucleobases, nucleosides, nucleotides and nucleic acids.
regulation of phosphate metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways involving phosphates.
regulation of metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways within a cell or an organism.
protein metabolic process
The chemical reactions and pathways involving a specific protein, rather than of proteins in general. Includes protein modification.
second-messenger-mediated signaling
A series of molecular signals in which an ion or small molecule is formed or released into the cytosol, thereby helping relay the signal within the cell.
cell cycle process
A cellular process that is involved in the progression of biochemical and morphological phases and events that occur in a cell during successive cell replication or nuclear replication events.
cell cycle phase
A cell cycle process comprising the steps by which a cell progresses through one of the biochemical and morphological phases and events that occur during successive cell replication or nuclear replication events.
regulation of anatomical structure morphogenesis
Any process that modulates the frequency, rate or extent of anatomical structure morphogenesis.
cell differentiation
The process whereby relatively unspecialized cells, e.g. embryonic or regenerative cells, acquire specialized structural and/or functional features that characterize the cells, tissues, or organs of the mature organism or some other relatively stable phase of the organism's life history. Differentiation includes the processes involved in commitment of a cell to a specific fate.
chromosome condensation
The progressive compaction of dispersed interphase chromatin into threadlike chromosomes prior to mitotic or meiotic nuclear division, or during apoptosis, in eukaryotic cells.
regulation of cell migration
Any process that modulates the frequency, rate or extent of cell migration.
regulation of cellular metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways by which individual cells transform chemical substances.
positive regulation of cellular metabolic process
Any process that activates or increases the frequency, rate or extent of the chemical reactions and pathways by which individual cells transform chemical substances.
regulation of cellular biosynthetic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways resulting in the formation of substances, carried out by individual cells.
regulation of protein modification process
Any process that modulates the frequency, rate or extent of the covalent alteration of one or more amino acid residues within a protein.
positive regulation of protein modification process
Any process that activates or increases the frequency, rate or extent of the covalent alteration of one or more amino acid residues within a protein.
regulation of cellular protein metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways involving a protein, occurring at the level of an individual cell.
positive regulation of cellular protein metabolic process
Any process that activates or increases the frequency, rate or extent of the chemical reactions and pathways involving a protein, occurring at the level of an individual cell.
multicellular organismal process
Any biological process, occurring at the level of a multicellular organism, pertinent to its function.
developmental process
A biological process whose specific outcome is the progression of an integrated living unit: an anatomical structure (which may be a subcellular structure, cell, tissue, or organ), or organism over time from an initial condition to a later condition.
regulation of localization
Any process that modulates the frequency, rate or extent of any process by which a cell, a substance, or a cellular entity is transported to, or maintained in, a specific location.
cellular response to stress
A change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus indicating the organism is under stress. The stress is usually, but not necessarily, exogenous (e.g. temperature, humidity, ionizing radiation).
cellular macromolecule biosynthetic process
The chemical reactions and pathways resulting in the formation of a macromolecule, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass, carried out by individual cells.
cellular biopolymer metabolic process
The chemical reactions and pathways involving biopolymers, long, repeating chains of monomers found in nature, such as polysaccharides and proteins, as carried out by individual cells.
cellular biopolymer biosynthetic process
The chemical reactions and pathways resulting in the formation of biopolymers, long, repeating chains of monomers found in nature, such as polysaccharides and proteins, as carried out by individual cells.
cellular response to DNA damage stimulus
A change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus indicating damage to its DNA from environmental insults or errors during metabolism.
locomotion
Self-propelled movement of a cell or organism from one location to another.
regulation of locomotion
Any process that modulates the frequency, rate or extent of locomotion of a cell or organism.
wound healing
The series of events that restore integrity to a damaged tissue, following an injury.
regulation of cell proliferation
Any process that modulates the frequency, rate or extent of cell proliferation.
regulation of phosphorylation
Any process that modulates the frequency, rate or extent of addition of phosphate groups into a molecule.
positive regulation of phosphorylation
Any process that activates or increases the frequency, rate or extent of addition of phosphate groups to a molecule.
macromolecule metabolic process
The chemical reactions and pathways involving macromolecules, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass.
biopolymer metabolic process
The chemical reactions and pathways involving biopolymers, long, repeating chains of monomers found in nature, such as polysaccharides and proteins.
biopolymer biosynthetic process
The chemical reactions and pathways resulting in the formation of biopolymers, long, repeating chains of monomers found in nature e.g. polysaccharides and proteins.
biopolymer modification
The covalent alteration of one or more monomeric units in a polypeptide, polynucleotide, polysaccharide, or other biological polymer, resulting in a change in its properties.
post-translational protein modification
The covalent alteration of one or more amino acids occurring in a protein after the protein has been completely translated and released from the ribosome.
cellular metabolic process
The chemical reactions and pathways by which individual cells transform chemical substances.
primary metabolic process
The chemical reactions and pathways involving those compounds which are formed as a part of the normal anabolic and catabolic processes. These processes take place in most, if not all, cells of the organism.
cellular biosynthetic process
The chemical reactions and pathways resulting in the formation of substances, carried out by individual cells.
cellular lipid metabolic process
The chemical reactions and pathways involving lipids, as carried out by individual cells.
cellular macromolecule metabolic process
The chemical reactions and pathways involving macromolecules, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass, as carried out by individual cells.
cellular protein metabolic process
The chemical reactions and pathways involving a specific protein, rather than of proteins in general, occurring at the level of an individual cell. Includes protein modification.
regulation of angiogenesis
Any process that modulates the frequency, rate or extent of angiogenesis.
positive regulation of angiogenesis
Any process that activates or increases angiogenesis.
positive regulation of cell cycle
Any process that activates or increases the rate or extent of progression through the cell cycle.
positive regulation of mitosis
Any process that activates or increases the frequency, rate or extent of mitosis.
positive regulation of phosphate metabolic process
Any process that activates or increases the frequency, rate or extent of the chemical reactions and pathways involving phosphates.
hormone secretion
The regulated release of hormones, substances with a specific regulatory effect on a particular organ or group of cells.
regulation of hormone secretion
Any process that modulates the frequency, rate or extent of the regulated release of a hormone from a cell or group of cells.
negative regulation of hormone secretion
Any process that stops, prevents or reduces the frequency, rate or extent of the regulated release of a hormone from a cell or group of cells.
organelle fission
The creation of two or more organelles by division of one organelle.
organ development
Development of a tissue or tissues that work together to perform a specific function or functions. Development pertains to the process whose specific outcome is the progression of a structure over time, from its formation to the mature structure. Organs are commonly observed as visibly distinct structures, but may also exist as loosely associated clusters of cells that work together to perform a specific function or functions.
blood vessel morphogenesis
The process by which the anatomical structures of blood vessels are generated and organized. Morphogenesis pertains to the creation of form. The blood vessel is the vasculature carrying blood.
positive regulation of biological process
Any process that activates or increases the frequency, rate or extent of a biological process. Biological processes are regulated by many means; examples include the control of gene expression, protein modification or interaction with a protein or substrate molecule.
negative regulation of biological process
Any process that stops, prevents or reduces the frequency, rate or extent of a biological process. Biological processes are regulated by many means; examples include the control of gene expression, protein modification or interaction with a protein or substrate molecule.
positive regulation of cellular process
Any process that activates or increases the frequency, rate or extent of a cellular process, any of those that are carried out at the cellular level, but are not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
negative regulation of cellular process
Any process that stops, prevents or reduces the frequency, rate or extent of a cellular process, any of those that are carried out at the cellular level, but are not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
anatomical structure formation
The process pertaining to the initial formation of an anatomical structure from unspecified parts. This process begins with the specific processes that contribute to the appearance of the discrete structure and ends when the structural rudiment is recognizable. An anatomical structure is any biological entity that occupies space and is distinguished from its surroundings. Anatomical structures can be macroscopic such as a carpel, or microscopic such as an acrosome.
system development
The process whose specific outcome is the progression of an organismal system over time, from its formation to the mature structure. A system is a regularly interacting or interdependent group of organs or tissues that work together to carry out a given biological process.
anatomical structure development
The biological process whose specific outcome is the progression of an anatomical structure from an initial condition to its mature state. This process begins with the formation of the structure and ends with the mature structure, whatever form that may be including its natural destruction. An anatomical structure is any biological entity that occupies space and is distinguished from its surroundings. Anatomical structures can be macroscopic such as a carpel, or microscopic such as an acrosome.
cellular developmental process
A biological process whose specific outcome is the progression of a cell over time from an initial condition to a later condition.
cell motility
Any process involved in the controlled movement of a cell that results in translocation of the cell from one place to another.
regulation of biological process
Any process that modulates the frequency, rate or extent of a biological process. Biological processes are regulated by many means; examples include the control of gene expression, protein modification or interaction with a protein or substrate molecule.
regulation of developmental process
Any process that modulates the frequency, rate or extent of development, the biological process whose specific outcome is the progression of a multicellular organism over time from an initial condition (e.g. a zygote, or a young adult) to a later condition (e.g. a multicellular animal or an aged adult).
regulation of cellular process
Any process that modulates the frequency, rate or extent of a cellular process, any of those that are carried out at the cellular level, but are not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
response to stimulus
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus.
leukocyte migration
The movement of leukocytes within or between different tissues and organs of the body.
regulation of secretion
Any process that modulates the frequency, rate or extent of the controlled release of a substance from a cell or group of cells.
negative regulation of secretion
Any process that stops, prevents or reduces the frequency, rate or extent of the controlled release of a substance from a cell or group of cells.
regulation of transport
Any process that modulates the frequency, rate or extent of the directed movement of substances (such as macromolecules, small molecules, ions) into, out of, within or between cells.
negative regulation of transport
Any process that stops, prevents or reduces the frequency, rate or extent of the directed movement of substances (such as macromolecules, small molecules, ions) into, out of, within or between cells.
regulation of DNA metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways involving DNA.
positive regulation of developmental process
Any process that activates or increases the rate or extent of development, the biological process whose specific outcome is the progression of an organism over time from an initial condition (e.g. a zygote, or a young adult) to a later condition (e.g. a multicellular animal or an aged adult).
regulation of phosphorus metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways involving phosphorus or compounds containing phosphorus.
localization
Any process by which a cell, a substance, or a cellular entity, such as a protein complex or organelle, is transported to, and/or maintained in a specific location.
establishment of localization
The directed movement of a cell, substance or cellular entity, such as a protein complex or organelle, to a specific location.
regulation of multicellular organismal process
Any process that modulates the frequency, rate or extent of a multicellular organismal process, the processes pertinent to the function of a multicellular organism above the cellular level; includes the integrated processes of tissues and organs.
regulation of protein metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways involving a protein.
positive regulation of protein metabolic process
Any process that activates or increases the frequency, rate or extent of the chemical reactions and pathways involving a protein.
regulation of cell motion
Any process that modulates the frequency, rate or extent of the movement of a cell.
cell division
The process resulting in the physical partitioning and separation of a cell into daughter cells.
S phase
A cell cycle process comprising the steps by which a cell progresses through S phase, the part of the cell cycle during which DNA synthesis takes place.
interphase
A cell cycle process comprising the steps by which a cell progresses through interphase, the stage of cell cycle between successive rounds of chromosome segregation. Canonically, interphase is the stage of the cell cycle during which the biochemical and physiologic functions of the cell are performed and replication of chromatin occurs.
interphase of mitotic cell cycle
A cell cycle process comprising the steps by which a cell progresses through interphase, the stage of cell cycle between successive rounds of mitosis. Canonically, interphase is the stage of the cell cycle during which the biochemical and physiologic functions of the cell are performed and replication of chromatin occurs.
localization of cell
Any process by which a cell is transported to, and/or maintained in, a specific location.
cellular response to stimulus
A change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus.
regulation of macromolecule metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways involving macromolecules, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass.
bone development
The process whose specific outcome is the progression of bone over time, from its formation to the mature structure. Bone is the hard skeletal connective tissue consisting of both mineral and cellular components.
biological regulation
Any process that modulates the frequency, rate or extent of any biological process, quality or function.
regulation of biological quality
Any process that modulates the frequency, rate or extent of a biological quality. A biological quality is a measurable attribute of an organism or part of an organism, such as size, mass, shape, color, etc.
all
This term is the most general term possible
cellular metabolic process
The chemical reactions and pathways by which individual cells transform chemical substances.
multicellular organismal development
The biological process whose specific outcome is the progression of a multicellular organism over time from an initial condition (e.g. a zygote or a young adult) to a later condition (e.g. a multicellular animal or an aged adult).
cellular developmental process
A biological process whose specific outcome is the progression of a cell over time from an initial condition to a later condition.
positive regulation of metabolic process
Any process that activates or increases the frequency, rate or extent of the chemical reactions and pathways within a cell or an organism.
positive regulation of cellular process
Any process that activates or increases the frequency, rate or extent of a cellular process, any of those that are carried out at the cellular level, but are not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
positive regulation of developmental process
Any process that activates or increases the rate or extent of development, the biological process whose specific outcome is the progression of an organism over time from an initial condition (e.g. a zygote, or a young adult) to a later condition (e.g. a multicellular animal or an aged adult).
negative regulation of cellular process
Any process that stops, prevents or reduces the frequency, rate or extent of a cellular process, any of those that are carried out at the cellular level, but are not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
regulation of metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways within a cell or an organism.
regulation of locomotion
Any process that modulates the frequency, rate or extent of locomotion of a cell or organism.
positive regulation of biological process
Any process that activates or increases the frequency, rate or extent of a biological process. Biological processes are regulated by many means; examples include the control of gene expression, protein modification or interaction with a protein or substrate molecule.
negative regulation of biological process
Any process that stops, prevents or reduces the frequency, rate or extent of a biological process. Biological processes are regulated by many means; examples include the control of gene expression, protein modification or interaction with a protein or substrate molecule.
regulation of developmental process
Any process that modulates the frequency, rate or extent of development, the biological process whose specific outcome is the progression of a multicellular organism over time from an initial condition (e.g. a zygote, or a young adult) to a later condition (e.g. a multicellular animal or an aged adult).
regulation of cellular process
Any process that modulates the frequency, rate or extent of a cellular process, any of those that are carried out at the cellular level, but are not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
regulation of multicellular organismal process
Any process that modulates the frequency, rate or extent of a multicellular organismal process, the processes pertinent to the function of a multicellular organism above the cellular level; includes the integrated processes of tissues and organs.
cellular response to stimulus
A change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus.
regulation of localization
Any process that modulates the frequency, rate or extent of any process by which a cell, a substance, or a cellular entity is transported to, or maintained in, a specific location.
establishment of localization
The directed movement of a cell, substance or cellular entity, such as a protein complex or organelle, to a specific location.
regulation of biological process
Any process that modulates the frequency, rate or extent of a biological process. Biological processes are regulated by many means; examples include the control of gene expression, protein modification or interaction with a protein or substrate molecule.
regulation of biosynthetic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways resulting in the formation of substances.
positive regulation of metabolic process
Any process that activates or increases the frequency, rate or extent of the chemical reactions and pathways within a cell or an organism.
macromolecule biosynthetic process
The chemical reactions and pathways resulting in the formation of a macromolecule, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass.
positive regulation of macromolecule metabolic process
Any process that increases the frequency, rate or extent of the chemical reactions and pathways involving macromolecules, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass.
regulation of macromolecule metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways involving macromolecules, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass.
regulation of cellular metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways by which individual cells transform chemical substances.
positive regulation of cellular metabolic process
Any process that activates or increases the frequency, rate or extent of the chemical reactions and pathways by which individual cells transform chemical substances.
cellular biosynthetic process
The chemical reactions and pathways resulting in the formation of substances, carried out by individual cells.
cellular macromolecule metabolic process
The chemical reactions and pathways involving macromolecules, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass, as carried out by individual cells.
nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
The chemical reactions and pathways involving nucleobases, nucleosides, nucleotides and nucleic acids.
cell motility
Any process involved in the controlled movement of a cell that results in translocation of the cell from one place to another.
cell cycle process
A cellular process that is involved in the progression of biochemical and morphological phases and events that occur in a cell during successive cell replication or nuclear replication events.
sister chromatid segregation
The process by which sister chromatids are organized and then physically separated and apportioned to two or more sets.
positive regulation of cell proliferation
Any process that activates or increases the rate or extent of cell proliferation.
positive regulation of cellular metabolic process
Any process that activates or increases the frequency, rate or extent of the chemical reactions and pathways by which individual cells transform chemical substances.
positive regulation of cell cycle
Any process that activates or increases the rate or extent of progression through the cell cycle.
regulation of cell cycle
Any process that modulates the rate or extent of progression through the cell cycle.
signal transduction
The cascade of processes by which a signal interacts with a receptor, causing a change in the level or activity of a second messenger or other downstream target, and ultimately effecting a change in the functioning of the cell.
regulation of cell communication
Any process that modulates the frequency, rate or extent of cell communication. Cell communication is the process that mediates interactions between a cell and its surroundings. Encompasses interactions such as signaling or attachment between one cell and another cell, between a cell and an extracellular matrix, or between a cell and any other aspect of its environment.
regulation of cellular metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways by which individual cells transform chemical substances.
regulation of cell proliferation
Any process that modulates the frequency, rate or extent of cell proliferation.
positive regulation of cellular process
Any process that activates or increases the frequency, rate or extent of a cellular process, any of those that are carried out at the cellular level, but are not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
negative regulation of cellular process
Any process that stops, prevents or reduces the frequency, rate or extent of a cellular process, any of those that are carried out at the cellular level, but are not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
regulation of cell motion
Any process that modulates the frequency, rate or extent of the movement of a cell.
anatomical structure formation
The process pertaining to the initial formation of an anatomical structure from unspecified parts. This process begins with the specific processes that contribute to the appearance of the discrete structure and ends when the structural rudiment is recognizable. An anatomical structure is any biological entity that occupies space and is distinguished from its surroundings. Anatomical structures can be macroscopic such as a carpel, or microscopic such as an acrosome.
anatomical structure morphogenesis
The process by which anatomical structures are generated and organized. Morphogenesis pertains to the creation of form.
system development
The process whose specific outcome is the progression of an organismal system over time, from its formation to the mature structure. A system is a regularly interacting or interdependent group of organs or tissues that work together to carry out a given biological process.
regulation of anatomical structure morphogenesis
Any process that modulates the frequency, rate or extent of anatomical structure morphogenesis.
positive regulation of developmental process
Any process that activates or increases the rate or extent of development, the biological process whose specific outcome is the progression of an organism over time from an initial condition (e.g. a zygote, or a young adult) to a later condition (e.g. a multicellular animal or an aged adult).
regulation of cell motion
Any process that modulates the frequency, rate or extent of the movement of a cell.
cellular response to stress
A change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus indicating the organism is under stress. The stress is usually, but not necessarily, exogenous (e.g. temperature, humidity, ionizing radiation).
response to wounding
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus indicating damage to the organism.
cell motion
Any process involved in the controlled movement of a cell.
regulation of transport
Any process that modulates the frequency, rate or extent of the directed movement of substances (such as macromolecules, small molecules, ions) into, out of, within or between cells.
negative regulation of transport
Any process that stops, prevents or reduces the frequency, rate or extent of the directed movement of substances (such as macromolecules, small molecules, ions) into, out of, within or between cells.
regulation of macromolecule biosynthetic process
Any process that modulates the rate, frequency or extent of the chemical reactions and pathways resulting in the formation of a macromolecule, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass.
regulation of cellular biosynthetic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways resulting in the formation of substances, carried out by individual cells.
cellular macromolecule biosynthetic process
The chemical reactions and pathways resulting in the formation of a macromolecule, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass, carried out by individual cells.
positive regulation of cellular metabolic process
Any process that activates or increases the frequency, rate or extent of the chemical reactions and pathways by which individual cells transform chemical substances.
regulation of cellular biosynthetic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways resulting in the formation of substances, carried out by individual cells.
regulation of macromolecule biosynthetic process
Any process that modulates the rate, frequency or extent of the chemical reactions and pathways resulting in the formation of a macromolecule, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass.
positive regulation of macromolecule metabolic process
Any process that increases the frequency, rate or extent of the chemical reactions and pathways involving macromolecules, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass.
protein metabolic process
The chemical reactions and pathways involving a specific protein, rather than of proteins in general. Includes protein modification.
biopolymer biosynthetic process
The chemical reactions and pathways resulting in the formation of biopolymers, long, repeating chains of monomers found in nature e.g. polysaccharides and proteins.
cellular macromolecule biosynthetic process
The chemical reactions and pathways resulting in the formation of a macromolecule, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass, carried out by individual cells.
cellular biopolymer metabolic process
The chemical reactions and pathways involving biopolymers, long, repeating chains of monomers found in nature, such as polysaccharides and proteins, as carried out by individual cells.
regulation of nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways involving nucleobases, nucleosides, nucleotides and nucleic acids.
positive regulation of phosphorus metabolic process
Any process that increases the frequency, rate or extent of the chemical reactions and pathways involving phosphorus or compounds containing phosphorus.
regulation of phosphorus metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways involving phosphorus or compounds containing phosphorus.
steroid metabolic process
The chemical reactions and pathways involving steroids, compounds with a 1,2,cyclopentanoperhydrophenanthrene nucleus.
cellular lipid metabolic process
The chemical reactions and pathways involving lipids, as carried out by individual cells.
regulation of protein metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways involving a protein.
positive regulation of protein metabolic process
Any process that activates or increases the frequency, rate or extent of the chemical reactions and pathways involving a protein.
negative regulation of hormone secretion
Any process that stops, prevents or reduces the frequency, rate or extent of the regulated release of a hormone from a cell or group of cells.
regulation of cell migration
Any process that modulates the frequency, rate or extent of cell migration.
regulation of mitotic cell cycle
Any process that modulates the rate or extent of progress through the mitotic cell cycle.
regulation of cell cycle process
Any process that modulates a cellular process that is involved in the progression of biochemical and morphological phases and events that occur in a cell during successive cell replication or nuclear replication events.
positive regulation of cell cycle
Any process that activates or increases the rate or extent of progression through the cell cycle.
generation of a signal involved in cell-cell signaling
The cellular process by which a physical entity or change in state, a signal, is created that originates in one cell and is used to transfer information to another cell. This process begins with the initial formation of the signal and ends with the mature form and placement of the signal.
regulation of hormone secretion
Any process that modulates the frequency, rate or extent of the regulated release of a hormone from a cell or group of cells.
positive regulation of cell proliferation
Any process that activates or increases the rate or extent of cell proliferation.
osteoblast differentiation
The process whereby a relatively unspecialized cell acquires the specialized features of an osteoblast, the mesodermal cell that gives rise to bone.
organ development
Development of a tissue or tissues that work together to perform a specific function or functions. Development pertains to the process whose specific outcome is the progression of a structure over time, from its formation to the mature structure. Organs are commonly observed as visibly distinct structures, but may also exist as loosely associated clusters of cells that work together to perform a specific function or functions.
positive regulation of angiogenesis
Any process that activates or increases angiogenesis.
regulation of angiogenesis
Any process that modulates the frequency, rate or extent of angiogenesis.
regulation of angiogenesis
Any process that modulates the frequency, rate or extent of angiogenesis.
positive regulation of angiogenesis
Any process that activates or increases angiogenesis.
organ morphogenesis
Morphogenesis of an organ. An organ is defined as a tissue or set of tissues that work together to perform a specific function or functions. Morphogenesis is the process by which anatomical structures are generated and organized. Organs are commonly observed as visibly distinct structures, but may also exist as loosely associated clusters of cells that work together to perform a specific function or functions.
ossification
The formation of bone or of a bony substance, or the conversion of fibrous tissue or of cartilage into bone or a bony substance.
regulation of cell migration
Any process that modulates the frequency, rate or extent of cell migration.
leukocyte migration
The movement of leukocytes within or between different tissues and organs of the body.
negative regulation of hormone secretion
Any process that stops, prevents or reduces the frequency, rate or extent of the regulated release of a hormone from a cell or group of cells.
negative regulation of transport
Any process that stops, prevents or reduces the frequency, rate or extent of the directed movement of substances (such as macromolecules, small molecules, ions) into, out of, within or between cells.
cellular response to DNA damage stimulus
A change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus indicating damage to its DNA from environmental insults or errors during metabolism.
hormone secretion
The regulated release of hormones, substances with a specific regulatory effect on a particular organ or group of cells.
regulation of secretion
Any process that modulates the frequency, rate or extent of the controlled release of a substance from a cell or group of cells.
negative regulation of secretion
Any process that stops, prevents or reduces the frequency, rate or extent of the controlled release of a substance from a cell or group of cells.
hormone secretion
The regulated release of hormones, substances with a specific regulatory effect on a particular organ or group of cells.
hormone transport
The directed movement of hormones into, out of, within or between cells.
cellular biopolymer biosynthetic process
The chemical reactions and pathways resulting in the formation of biopolymers, long, repeating chains of monomers found in nature, such as polysaccharides and proteins, as carried out by individual cells.
regulation of DNA replication
Any process that modulates the frequency, rate or extent of DNA replication.
sterol metabolic process
The chemical reactions and pathways involving sterols, steroids with one or more hydroxyl groups and a hydrocarbon side-chain in the molecule.
positive regulation of cellular protein metabolic process
Any process that activates or increases the frequency, rate or extent of the chemical reactions and pathways involving a protein, occurring at the level of an individual cell.
regulation of DNA metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways involving DNA.
positive regulation of cellular protein metabolic process
Any process that activates or increases the frequency, rate or extent of the chemical reactions and pathways involving a protein, occurring at the level of an individual cell.
positive regulation of phosphorus metabolic process
Any process that increases the frequency, rate or extent of the chemical reactions and pathways involving phosphorus or compounds containing phosphorus.
regulation of DNA replication
Any process that modulates the frequency, rate or extent of DNA replication.
regulation of cellular protein metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways involving a protein, occurring at the level of an individual cell.
positive regulation of protein metabolic process
Any process that activates or increases the frequency, rate or extent of the chemical reactions and pathways involving a protein.
DNA metabolic process
The chemical reactions and pathways involving DNA, deoxyribonucleic acid, one of the two main types of nucleic acid, consisting of a long, unbranched macromolecule formed from one, or more commonly, two, strands of linked deoxyribonucleotides.
cellular biopolymer biosynthetic process
The chemical reactions and pathways resulting in the formation of biopolymers, long, repeating chains of monomers found in nature, such as polysaccharides and proteins, as carried out by individual cells.
cellular protein metabolic process
The chemical reactions and pathways involving a specific protein, rather than of proteins in general, occurring at the level of an individual cell. Includes protein modification.
regulation of DNA metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways involving DNA.
regulation of phosphate metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways involving phosphates.
positive regulation of phosphate metabolic process
Any process that activates or increases the frequency, rate or extent of the chemical reactions and pathways involving phosphates.
protein modification process
The covalent alteration of one or more amino acids occurring in proteins, peptides and nascent polypeptides (co-translational, post-translational modifications). Includes the modification of charged tRNAs that are destined to occur in a protein (pre-translation modification).
regulation of cellular protein metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways involving a protein, occurring at the level of an individual cell.
positive regulation of cellular protein metabolic process
Any process that activates or increases the frequency, rate or extent of the chemical reactions and pathways involving a protein, occurring at the level of an individual cell.
mitotic chromosome condensation
The cell cycle process whereby chromatin structure is compacted prior to mitosis in eukaryotic cells.
negative regulation of hormone secretion
Any process that stops, prevents or reduces the frequency, rate or extent of the regulated release of a hormone from a cell or group of cells.
mitosis
A cell cycle process comprising the steps by which the nucleus of a eukaryotic cell divides; the process involves condensation of chromosomal DNA into a highly compacted form. Canonically, mitosis produces two daughter nuclei whose chromosome complement is identical to that of the mother cell.
regulation of mitosis
Any process that modulates the frequency, rate or extent of mitosis.
mitotic chromosome condensation
The cell cycle process whereby chromatin structure is compacted prior to mitosis in eukaryotic cells.
sister chromatid segregation
The process by which sister chromatids are organized and then physically separated and apportioned to two or more sets.
chromosome condensation
The progressive compaction of dispersed interphase chromatin into threadlike chromosomes prior to mitotic or meiotic nuclear division, or during apoptosis, in eukaryotic cells.
microtubule cytoskeleton organization
A process that is carried out at the cellular level which results in the formation, arrangement of constituent parts, or disassembly of cytoskeletal structures comprising microtubules and their associated proteins.
positive regulation of mitosis
Any process that activates or increases the frequency, rate or extent of mitosis.
M phase of mitotic cell cycle
A cell cycle process comprising the steps by which a cell progresses through M phase, the part of the mitotic cell cycle during which mitosis takes place.
mitotic sister chromatid segregation
The cell cycle process whereby replicated homologous chromosomes are organized and then physically separated and apportioned to two sets during the mitotic cell cycle. Each replicated chromosome, composed of two sister chromatids, aligns at the cell equator, paired with its homologous partner. One homolog of each morphologic type goes into each of the resulting chromosome sets.
regulation of mitosis
Any process that modulates the frequency, rate or extent of mitosis.
positive regulation of mitosis
Any process that activates or increases the frequency, rate or extent of mitosis.
S phase
A cell cycle process comprising the steps by which a cell progresses through S phase, the part of the cell cycle during which DNA synthesis takes place.
interphase of mitotic cell cycle
A cell cycle process comprising the steps by which a cell progresses through interphase, the stage of cell cycle between successive rounds of mitosis. Canonically, interphase is the stage of the cell cycle during which the biochemical and physiologic functions of the cell are performed and replication of chromatin occurs.
DNA repair
The process of restoring DNA after damage. Genomes are subject to damage by chemical and physical agents in the environment (e.g. UV and ionizing radiations, chemical mutagens, fungal and bacterial toxins, etc.) and by free radicals or alkylating agents endogenously generated in metabolism. DNA is also damaged because of errors during its replication. A variety of different DNA repair pathways have been reported that include direct reversal, base excision repair, nucleotide excision repair, photoreactivation, bypass, double-strand break repair pathway, and mismatch repair pathway.
bone development
The process whose specific outcome is the progression of bone over time, from its formation to the mature structure. Bone is the hard skeletal connective tissue consisting of both mineral and cellular components.
angiogenesis
Blood vessel formation when new vessels emerge from the proliferation of pre-existing blood vessels.
regulation of hormone secretion
Any process that modulates the frequency, rate or extent of the regulated release of a hormone from a cell or group of cells.
negative regulation of secretion
Any process that stops, prevents or reduces the frequency, rate or extent of the controlled release of a substance from a cell or group of cells.
DNA replication
The process whereby new strands of DNA are synthesized. The template for replication can either be an existing DNA molecule or RNA.
positive regulation of protein modification process
Any process that activates or increases the frequency, rate or extent of the covalent alteration of one or more amino acid residues within a protein.
positive regulation of phosphate metabolic process
Any process that activates or increases the frequency, rate or extent of the chemical reactions and pathways involving phosphates.
regulation of protein modification process
Any process that modulates the frequency, rate or extent of the covalent alteration of one or more amino acid residues within a protein.
positive regulation of protein modification process
Any process that activates or increases the frequency, rate or extent of the covalent alteration of one or more amino acid residues within a protein.
regulation of DNA replication
Any process that modulates the frequency, rate or extent of DNA replication.
DNA synthesis during DNA repair
Synthesis of DNA that proceeds from the broken 3' single-strand DNA end uses the homologous intact duplex as the template.
recombinational repair
The repair of damaged DNA that involves the exchange, reciprocal or nonreciprocal, of genetic material between the broken DNA molecule and a homologous region of DNA.
regulation of phosphorylation
Any process that modulates the frequency, rate or extent of addition of phosphate groups into a molecule.
positive regulation of phosphorylation
Any process that activates or increases the frequency, rate or extent of addition of phosphate groups to a molecule.
mitosis
A cell cycle process comprising the steps by which the nucleus of a eukaryotic cell divides; the process involves condensation of chromosomal DNA into a highly compacted form. Canonically, mitosis produces two daughter nuclei whose chromosome complement is identical to that of the mother cell.
blood vessel morphogenesis
The process by which the anatomical structures of blood vessels are generated and organized. Morphogenesis pertains to the creation of form. The blood vessel is the vasculature carrying blood.
regulation of protein amino acid phosphorylation
Any process that modulates the frequency, rate or extent of addition of phosphate groups into an amino acid in a protein.
positive regulation of phosphorylation
Any process that activates or increases the frequency, rate or extent of addition of phosphate groups to a molecule.
protein amino acid phosphorylation
The process of introducing a phosphate group on to a protein.
DNA replication initiation
The process by which DNA replication is started; this involves the separation of a stretch of the DNA double helix, the recruitment of DNA polymerases and the initiation of polymerase action.
nucleotide-excision repair, DNA gap filling
Repair of the gap in the DNA helix by DNA polymerase and DNA ligase after the portion of the strand containing the lesion has been removed by pyrimidine-dimer repair enzymes.
double-strand break repair via homologous recombination
The error-free repair of a double-strand break in DNA in which the broken DNA molecule is repaired using homologous sequences. A strand in the broken DNA searches for a homologous region in an intact chromosome to serve as the template for DNA synthesis. The restoration of two intact DNA molecules results in the exchange, reciprocal or nonreciprocal, of genetic material between the intact DNA molecule and the broken DNA molecule.
regulation of protein amino acid phosphorylation
Any process that modulates the frequency, rate or extent of addition of phosphate groups into an amino acid in a protein.

nuclear chromosome
A chromosome found in the nucleus of a eukaryotic cell.
condensed chromosome
A highly compacted molecule of DNA and associated proteins resulting in a cytologically distinct structure.
intracellular
The living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
chromosome, centromeric region
The region of a chromosome that includes the centromere and associated proteins. In monocentric chromosomes, this region corresponds to a single area of the chromosome, whereas in holocentric chromosomes, it is evenly distributed along the chromosome.
kinetochore
A multisubunit complex that is located at the centromeric region of DNA and provides an attachment point for the spindle microtubules.
condensed chromosome kinetochore
A multisubunit complex that is located at the centromeric region of a condensed chromosome and provides an attachment point for the spindle microtubules.
condensed chromosome, centromeric region
The region of a condensed chromosome that includes the centromere and associated proteins, including the kinetochore. In monocentric chromosomes, this region corresponds to a single area of the chromosome, whereas in holocentric chromosomes, it is evenly distributed along the chromosome.
condensin complex
A multisubunit protein complex that plays a central role in chromosome condensation.
extracellular region
The space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers the host cell environment outside an intracellular parasite.
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
extracellular space
That part of a multicellular organism outside the cells proper, usually taken to be outside the plasma membranes, and occupied by fluid.
cell
The basic structural and functional unit of all organisms. Includes the plasma membrane and any external encapsulating structures such as the cell wall and cell envelope.
nucleus
A membrane-bounded organelle of eukaryotic cells in which chromosomes are housed and replicated. In most cells, the nucleus contains all of the cell's chromosomes except the organellar chromosomes, and is the site of RNA synthesis and processing. In some species, or in specialized cell types, RNA metabolism or DNA replication may be absent.
replication fork
The Y-shaped region of a replicating DNA molecule, resulting from the separation of the DNA strands and in which the synthesis of new strands takes place. Also includes associated protein complexes.
alpha DNA polymerase:primase complex
A complex of four polypeptides, comprising large and small DNA polymerase alpha subunits and two primase subunits, which catalyzes the synthesis of an RNA primer on the lagging strand of replicating DNA; the smaller of the two primase subunits alone can catalyze oligoribonucleotide synthesis.
chromosome
A structure composed of a very long molecule of DNA and associated proteins (e.g. histones) that carries hereditary information.
spindle
The array of microtubules and associated molecules that forms between opposite poles of a eukaryotic cell during mitosis or meiosis and serves to move the duplicated chromosomes apart.
cytoskeleton
Any of the various filamentous elements that form the internal framework of cells, and typically remain after treatment of the cells with mild detergent to remove membrane constituents and soluble components of the cytoplasm. The term embraces intermediate filaments, microfilaments, microtubules, the microtrabecular lattice, and other structures characterized by a polymeric filamentous nature and long-range order within the cell. The various elements of the cytoskeleton not only serve in the maintenance of cellular shape but also have roles in other cellular functions, including cellular movement, cell division, endocytosis, and movement of organelles.
microtubule cytoskeleton
The part of the cytoskeleton (the internal framework of a cell) composed of microtubules and associated proteins.
replisome
A multi-component enzymatic machine at the replication fork which mediates DNA replication. Includes DNA primase, one or more DNA polymerases, DNA helicases, and other proteins.
membrane-enclosed lumen
The enclosed volume within a sealed membrane or between two sealed membranes. Encompasses the volume enclosed by the membranes of a particular organelle, e.g. endoplasmic reticulum lumen, or the space between the two lipid bilayers of a double membrane surrounding an organelle, e.g. nuclear envelope lumen.
nuclear lumen
The volume enclosed by the nuclear inner membrane.
macromolecular complex
A stable assembly of two or more macromolecules, i.e. proteins, nucleic acids, carbohydrates or lipids, in which the constituent parts function together.
protein-DNA complex
A macromolecular complex containing both protein and DNA molecules.
organelle
Organized structure of distinctive morphology and function. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton. Excludes the plasma membrane.
membrane-bounded organelle
Organized structure of distinctive morphology and function, bounded by a single or double lipid bilayer membrane. Includes the nucleus, mitochondria, plastids, vacuoles, and vesicles. Excludes the plasma membrane.
non-membrane-bounded organelle
Organized structure of distinctive morphology and function, not bounded by a lipid bilayer membrane. Includes ribosomes, the cytoskeleton and chromosomes.
intracellular organelle
Organized structure of distinctive morphology and function, occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton. Excludes the plasma membrane.
intracellular membrane-bounded organelle
Organized structure of distinctive morphology and function, bounded by a single or double lipid bilayer membrane and occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, and vesicles. Excludes the plasma membrane.
intracellular non-membrane-bounded organelle
Organized structure of distinctive morphology and function, not bounded by a lipid bilayer membrane and occurring within the cell. Includes ribosomes, the cytoskeleton and chromosomes.
organelle lumen
The internal volume enclosed by the membranes of a particular organelle; includes the volume enclosed by a single organelle membrane, e.g. endoplasmic reticulum lumen, or the volume enclosed by the innermost of the two lipid bilayers of an organelle envelope, e.g. nuclear lumen.
protein complex
Any macromolecular complex composed of two or more polypeptide subunits, which may or may not be identical. Protein complexes may have other associated non-protein prosthetic groups, such as nucleotides, metal ions or carbohydrate groups.
nuclear replication fork
The Y-shaped region of a nuclear replicating DNA molecule, resulting from the separation of the DNA strands and in which the synthesis of new strands takes place. Also includes associated protein complexes.
nuclear replisome
A multi-component enzymatic machine at the nuclear replication fork, which mediates DNA replication. Includes DNA primase, one or more DNA polymerases, DNA helicases, and other proteins.
extracellular region part
Any constituent part of the extracellular region, the space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers constituent parts of the host cell environment outside an intracellular parasite.
organelle part
Any constituent part of an organelle, an organized structure of distinctive morphology and function. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton, but excludes the plasma membrane.
intracellular part
Any constituent part of the living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
chromosomal part
Any constituent part of a chromosome, a structure composed of a very long molecule of DNA and associated proteins (e.g. histones) that carries hereditary information.
nuclear part
Any constituent part of the nucleus, a membrane-bounded organelle of eukaryotic cells in which chromosomes are housed and replicated.
cytoskeletal part
Any constituent part of the cytoskeleton, a cellular scaffolding or skeleton that maintains cell shape, enables some cell motion (using structures such as flagella and cilia), and plays important roles in both intra-cellular transport (e.g. the movement of vesicles and organelles) and cellular division. Includes constituent parts of intermediate filaments, microfilaments, microtubules, and the microtrabecular lattice.
intracellular organelle part
A constituent part of an intracellular organelle, an organized structure of distinctive morphology and function, occurring within the cell. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton but excludes the plasma membrane.
nuclear chromosome part
Any constituent part of a nuclear chromosome, a chromosome found in the nucleus of a eukaryotic cell.
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
intracellular organelle lumen
An organelle lumen that is part of an intracellular organelle.
all
This term is the most general term possible
extracellular region part
Any constituent part of the extracellular region, the space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers constituent parts of the host cell environment outside an intracellular parasite.
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
organelle part
Any constituent part of an organelle, an organized structure of distinctive morphology and function. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton, but excludes the plasma membrane.
organelle lumen
The internal volume enclosed by the membranes of a particular organelle; includes the volume enclosed by a single organelle membrane, e.g. endoplasmic reticulum lumen, or the volume enclosed by the innermost of the two lipid bilayers of an organelle envelope, e.g. nuclear lumen.
intracellular membrane-bounded organelle
Organized structure of distinctive morphology and function, bounded by a single or double lipid bilayer membrane and occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, and vesicles. Excludes the plasma membrane.
intracellular non-membrane-bounded organelle
Organized structure of distinctive morphology and function, not bounded by a lipid bilayer membrane and occurring within the cell. Includes ribosomes, the cytoskeleton and chromosomes.
intracellular organelle part
A constituent part of an intracellular organelle, an organized structure of distinctive morphology and function, occurring within the cell. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton but excludes the plasma membrane.
intracellular organelle lumen
An organelle lumen that is part of an intracellular organelle.
intracellular part
Any constituent part of the living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
intracellular organelle
Organized structure of distinctive morphology and function, occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton. Excludes the plasma membrane.
intracellular organelle part
A constituent part of an intracellular organelle, an organized structure of distinctive morphology and function, occurring within the cell. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton but excludes the plasma membrane.
kinetochore
A multisubunit complex that is located at the centromeric region of DNA and provides an attachment point for the spindle microtubules.
kinetochore
A multisubunit complex that is located at the centromeric region of DNA and provides an attachment point for the spindle microtubules.
condensin complex
A multisubunit protein complex that plays a central role in chromosome condensation.
replisome
A multi-component enzymatic machine at the replication fork which mediates DNA replication. Includes DNA primase, one or more DNA polymerases, DNA helicases, and other proteins.
nuclear lumen
The volume enclosed by the nuclear inner membrane.
nuclear chromosome part
Any constituent part of a nuclear chromosome, a chromosome found in the nucleus of a eukaryotic cell.
spindle
The array of microtubules and associated molecules that forms between opposite poles of a eukaryotic cell during mitosis or meiosis and serves to move the duplicated chromosomes apart.
alpha DNA polymerase:primase complex
A complex of four polypeptides, comprising large and small DNA polymerase alpha subunits and two primase subunits, which catalyzes the synthesis of an RNA primer on the lagging strand of replicating DNA; the smaller of the two primase subunits alone can catalyze oligoribonucleotide synthesis.
nuclear part
Any constituent part of the nucleus, a membrane-bounded organelle of eukaryotic cells in which chromosomes are housed and replicated.
nuclear chromosome
A chromosome found in the nucleus of a eukaryotic cell.
chromosomal part
Any constituent part of a chromosome, a structure composed of a very long molecule of DNA and associated proteins (e.g. histones) that carries hereditary information.
cytoskeletal part
Any constituent part of the cytoskeleton, a cellular scaffolding or skeleton that maintains cell shape, enables some cell motion (using structures such as flagella and cilia), and plays important roles in both intra-cellular transport (e.g. the movement of vesicles and organelles) and cellular division. Includes constituent parts of intermediate filaments, microfilaments, microtubules, and the microtrabecular lattice.
kinetochore
A multisubunit complex that is located at the centromeric region of DNA and provides an attachment point for the spindle microtubules.
replisome
A multi-component enzymatic machine at the replication fork which mediates DNA replication. Includes DNA primase, one or more DNA polymerases, DNA helicases, and other proteins.
alpha DNA polymerase:primase complex
A complex of four polypeptides, comprising large and small DNA polymerase alpha subunits and two primase subunits, which catalyzes the synthesis of an RNA primer on the lagging strand of replicating DNA; the smaller of the two primase subunits alone can catalyze oligoribonucleotide synthesis.
nuclear replication fork
The Y-shaped region of a nuclear replicating DNA molecule, resulting from the separation of the DNA strands and in which the synthesis of new strands takes place. Also includes associated protein complexes.
nuclear replisome
A multi-component enzymatic machine at the nuclear replication fork, which mediates DNA replication. Includes DNA primase, one or more DNA polymerases, DNA helicases, and other proteins.
nuclear chromosome part
Any constituent part of a nuclear chromosome, a chromosome found in the nucleus of a eukaryotic cell.
condensed chromosome, centromeric region
The region of a condensed chromosome that includes the centromere and associated proteins, including the kinetochore. In monocentric chromosomes, this region corresponds to a single area of the chromosome, whereas in holocentric chromosomes, it is evenly distributed along the chromosome.
condensin complex
A multisubunit protein complex that plays a central role in chromosome condensation.
spindle
The array of microtubules and associated molecules that forms between opposite poles of a eukaryotic cell during mitosis or meiosis and serves to move the duplicated chromosomes apart.
condensed chromosome kinetochore
A multisubunit complex that is located at the centromeric region of a condensed chromosome and provides an attachment point for the spindle microtubules.
nuclear replisome
A multi-component enzymatic machine at the nuclear replication fork, which mediates DNA replication. Includes DNA primase, one or more DNA polymerases, DNA helicases, and other proteins.

protein binding
Interacting selectively with any protein or protein complex (a complex of two or more proteins that may include other nonprotein molecules).
molecular_function
Elemental activities, such as catalysis or binding, describing the actions of a gene product at the molecular level. A given gene product may exhibit one or more molecular functions.
nucleic acid binding
Interacting selectively with any nucleic acid.
DNA binding
Interacting selectively with DNA (deoxyribonucleic acid).
catalytic activity
Catalysis of a biochemical reaction at physiological temperatures. In biologically catalyzed reactions, the reactants are known as substrates, and the catalysts are naturally occurring macromolecular substances known as enzymes. Enzymes possess specific binding sites for substrates, and are usually composed wholly or largely of protein, but RNA that has catalytic activity (ribozyme) is often also regarded as enzymatic.
DNA-directed DNA polymerase activity
Catalysis of the reaction: deoxynucleoside triphosphate + DNA(n) = diphosphate + DNA(n+1); the synthesis of DNA from deoxyribonucleotide triphosphates in the presence of a DNA template and primer.
receptor binding
Interacting selectively with one or more specific sites on a receptor molecule, a macromolecule that undergoes combination with a hormone, neurotransmitter, drug or intracellular messenger to initiate a change in cell function.
cytokine activity
Functions to control the survival, growth, differentiation and effector function of tissues and cells.
binding
The selective, often stoichiometric, interaction of a molecule with one or more specific sites on another molecule.
growth factor activity
The function that stimulates a cell to grow or proliferate. Most growth factors have other actions besides the induction of cell growth or proliferation.
transferase activity
Catalysis of the transfer of a group, e.g. a methyl group, glycosyl group, acyl group, phosphorus-containing, or other groups, from one compound (generally regarded as the donor) to another compound (generally regarded as the acceptor). Transferase is the systematic name for any enzyme of EC class 2.
transferase activity, transferring phosphorus-containing groups
Catalysis of the transfer of a phosphorus-containing group from one compound (donor) to another (acceptor).
nucleotidyltransferase activity
Catalysis of the transfer of a nucleotidyl group to a reactant.
DNA polymerase activity
Catalysis of the reaction: deoxynucleoside triphosphate + DNA(n) = diphosphate + DNA(n+1); the synthesis of DNA from deoxyribonucleotide triphosphates in the presence of a nucleic acid template and primer.
all
This term is the most general term possible
| Id | Pvalue | ExpCount | Count | Size | Term |
|---|---|---|---|---|---|
| 03030 | 9.192e-10 | 2.191 | 17 | 33 | DNA replication |
| 04110 | 2.118e-06 | 6.507 | 24 | 98 | Cell cycle |
| 04060 | 2.244e-02 | 8.101 | 19 | 122 | Cytokine-cytokine receptor interaction |
| 03430 | 2.511e-02 | 1.461 | 7 | 22 | Mismatch repair |
| 00100 | 3.225e-02 | 1.527 | 7 | 23 | Biosynthesis of steroids |
| 03410 | 3.401e-02 | 1.992 | 8 | 30 | Base excision repair |
| 04010 | 4.998e-02 | 12.35 | 24 | 186 | MAPK signaling pathway |
ABCA5ATP-binding cassette, sub-family A (ABC1), member 5 (213353_at), score: 0.41 ADIPOR2adiponectin receptor 2 (201346_at), score: 0.58 ADMadrenomedullin (202912_at), score: -0.78 AGFG2ArfGAP with FG repeats 2 (222126_at), score: -0.79 AJAP1adherens junctions associated protein 1 (206460_at), score: 0.63 ALDH6A1aldehyde dehydrogenase 6 family, member A1 (221589_s_at), score: 0.4 ALDOCaldolase C, fructose-bisphosphate (202022_at), score: 0.44 ANGPTL4angiopoietin-like 4 (221009_s_at), score: 1 ANK2ankyrin 2, neuronal (202920_at), score: 0.63 AQP3aquaporin 3 (Gill blood group) (39248_at), score: 0.72 AREGamphiregulin (205239_at), score: 0.56 ARHGAP11ARho GTPase activating protein 11A (204492_at), score: -0.77 ARID5AAT rich interactive domain 5A (MRF1-like) (213138_at), score: 0.57 ASAP2ArfGAP with SH3 domain, ankyrin repeat and PH domain 2 (206414_s_at), score: 0.55 ASF1BASF1 anti-silencing function 1 homolog B (S. cerevisiae) (218115_at), score: -0.78 ASPMasp (abnormal spindle) homolog, microcephaly associated (Drosophila) (219918_s_at), score: -0.82 ATF3activating transcription factor 3 (202672_s_at), score: 0.46 ATP13A3ATPase type 13A3 (219558_at), score: 0.69 ATP2A2ATPase, Ca++ transporting, cardiac muscle, slow twitch 2 (212361_s_at), score: 0.45 ATP6V0A1ATPase, H+ transporting, lysosomal V0 subunit a1 (205095_s_at), score: 0.43 AURKAaurora kinase A (208079_s_at), score: -0.77 AURKBaurora kinase B (209464_at), score: -0.85 B3GNT2UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 2 (219326_s_at), score: 0.72 BBS1Bardet-Biedl syndrome 1 (218471_s_at), score: 0.43 BCKDHBbranched chain keto acid dehydrogenase E1, beta polypeptide (210653_s_at), score: 0.44 BCL7AB-cell CLL/lymphoma 7A (203795_s_at), score: 0.55 BHLHE40basic helix-loop-helix family, member e40 (201170_s_at), score: 0.47 BIRC5baculoviral IAP repeat-containing 5 (202095_s_at), score: -0.84 BLMBloom syndrome (205733_at), score: -0.84 BMP2bone morphogenetic protein 2 (205289_at), score: 0.72 BMP6bone morphogenetic protein 6 (206176_at), score: 0.54 BMPR2bone morphogenetic protein receptor, type II (serine/threonine kinase) (210214_s_at), score: 0.66 BRCA1breast cancer 1, early onset (204531_s_at), score: -0.75 BRCA2breast cancer 2, early onset (208368_s_at), score: -0.85 BUB1budding uninhibited by benzimidazoles 1 homolog (yeast) (209642_at), score: -0.79 BUB1Bbudding uninhibited by benzimidazoles 1 homolog beta (yeast) (203755_at), score: -0.8 C11orf75chromosome 11 open reading frame 75 (219806_s_at), score: 0.42 C12orf48chromosome 12 open reading frame 48 (220060_s_at), score: -0.76 C17orf91chromosome 17 open reading frame 91 (214696_at), score: 0.85 C18orf24chromosome 18 open reading frame 24 (217640_x_at), score: -0.82 C19orf28chromosome 19 open reading frame 28 (220178_at), score: 0.48 C1Rcomplement component 1, r subcomponent (212067_s_at), score: 0.55 C20orf27chromosome 20 open reading frame 27 (50314_i_at), score: -0.8 C21orf45chromosome 21 open reading frame 45 (219004_s_at), score: -0.91 C3orf52chromosome 3 open reading frame 52 (219474_at), score: 0.46 C4orf18chromosome 4 open reading frame 18 (219872_at), score: 0.54 C6orf145chromosome 6 open reading frame 145 (212923_s_at), score: 0.48 CASP1caspase 1, apoptosis-related cysteine peptidase (interleukin 1, beta, convertase) (211368_s_at), score: 0.44 CASP4caspase 4, apoptosis-related cysteine peptidase (209310_s_at), score: 0.4 CBFBcore-binding factor, beta subunit (206788_s_at), score: 0.43 CBLBCas-Br-M (murine) ecotropic retroviral transforming sequence b (209682_at), score: 0.53 CBR3carbonyl reductase 3 (205379_at), score: -0.77 CCDC93coiled-coil domain containing 93 (209688_s_at), score: 0.39 CCL2chemokine (C-C motif) ligand 2 (216598_s_at), score: 0.62 CCNA2cyclin A2 (203418_at), score: -0.77 CCNB2cyclin B2 (202705_at), score: -0.81 CCNE2cyclin E2 (205034_at), score: -0.82 CCNJcyclin J (219470_x_at), score: 0.5 CD302CD302 molecule (203799_at), score: 0.57 CD55CD55 molecule, decay accelerating factor for complement (Cromer blood group) (201925_s_at), score: 0.61 CDC14BCDC14 cell division cycle 14 homolog B (S. cerevisiae) (208022_s_at), score: 0.42 CDC20cell division cycle 20 homolog (S. cerevisiae) (202870_s_at), score: -0.81 CDC45LCDC45 cell division cycle 45-like (S. cerevisiae) (204126_s_at), score: -0.81 CDC6cell division cycle 6 homolog (S. cerevisiae) (203967_at), score: -0.82 CDC7cell division cycle 7 homolog (S. cerevisiae) (204510_at), score: -0.79 CDCA3cell division cycle associated 3 (221436_s_at), score: -0.78 CDCA8cell division cycle associated 8 (221520_s_at), score: -0.81 CDK2cyclin-dependent kinase 2 (204252_at), score: -0.87 CDT1chromatin licensing and DNA replication factor 1 (209832_s_at), score: -0.84 CDV3CDV3 homolog (mouse) (213548_s_at), score: 0.4 CDYLchromodomain protein, Y-like (203100_s_at), score: 0.59 CENPFcentromere protein F, 350/400ka (mitosin) (207828_s_at), score: -0.79 CENPMcentromere protein M (218741_at), score: -0.86 CEP55centrosomal protein 55kDa (218542_at), score: -0.79 CHAF1Achromatin assembly factor 1, subunit A (p150) (214426_x_at), score: -0.85 CHAF1Bchromatin assembly factor 1, subunit B (p60) (204775_at), score: -0.8 CHIC2cysteine-rich hydrophobic domain 2 (219492_at), score: 0.47 CHMP1Bchromatin modifying protein 1B (218178_s_at), score: 0.55 CKAP2cytoskeleton associated protein 2 (218252_at), score: -0.87 CLCF1cardiotrophin-like cytokine factor 1 (219500_at), score: 0.48 CNTLNcentlein, centrosomal protein (220095_at), score: -0.77 COL14A1collagen, type XIV, alpha 1 (212865_s_at), score: 0.53 COL15A1collagen, type XV, alpha 1 (203477_at), score: 0.71 COQ10Bcoenzyme Q10 homolog B (S. cerevisiae) (219397_at), score: 0.45 CORINcorin, serine peptidase (220356_at), score: 0.63 CPA3carboxypeptidase A3 (mast cell) (205624_at), score: 0.59 CREG1cellular repressor of E1A-stimulated genes 1 (201200_at), score: 0.55 CRISPLD2cysteine-rich secretory protein LCCL domain containing 2 (221541_at), score: 0.5 CSGALNACT2chondroitin sulfate N-acetylgalactosaminyltransferase 2 (222235_s_at), score: 0.57 CSRNP2cysteine-serine-rich nuclear protein 2 (221260_s_at), score: 0.47 CTSOcathepsin O (203758_at), score: 0.48 CXCL3chemokine (C-X-C motif) ligand 3 (207850_at), score: 0.59 CXCR7chemokine (C-X-C motif) receptor 7 (212977_at), score: 0.52 CYP51A1cytochrome P450, family 51, subfamily A, polypeptide 1 (216607_s_at), score: 0.63 DACT1dapper, antagonist of beta-catenin, homolog 1 (Xenopus laevis) (219179_at), score: 0.44 DCP1ADCP1 decapping enzyme homolog A (S. cerevisiae) (218508_at), score: 0.51 DDX11DEAD/H (Asp-Glu-Ala-Asp/His) box polypeptide 11 (CHL1-like helicase homolog, S. cerevisiae) (208149_x_at), score: -0.88 DDX12DEAD/H (Asp-Glu-Ala-Asp/His) box polypeptide 12 (CHL1-like helicase homolog, S. cerevisiae) (213378_s_at), score: -0.83 DDX3YDEAD (Asp-Glu-Ala-Asp) box polypeptide 3, Y-linked (205000_at), score: 0.53 DEKDEK oncogene (200934_at), score: -0.76 DENND3DENN/MADD domain containing 3 (212975_at), score: 0.45 DEPDC1DEP domain containing 1 (220295_x_at), score: -0.86 DGKDdiacylglycerol kinase, delta 130kDa (208072_s_at), score: 0.41 DHCR77-dehydrocholesterol reductase (201790_s_at), score: 0.57 DHFRdihydrofolate reductase (202533_s_at), score: -0.76 DKK2dickkopf homolog 2 (Xenopus laevis) (219908_at), score: 0.46 DLEU2deleted in lymphocytic leukemia 2 (non-protein coding) (216870_x_at), score: -0.77 DLEU2Ldeleted in lymphocytic leukemia 2-like (215629_s_at), score: -0.79 DLGAP5discs, large (Drosophila) homolog-associated protein 5 (203764_at), score: -0.81 DNASE2deoxyribonuclease II, lysosomal (214992_s_at), score: 0.52 DOCK9dedicator of cytokinesis 9 (212538_at), score: 0.58 DPP4dipeptidyl-peptidase 4 (211478_s_at), score: 0.49 DRAMdamage-regulated autophagy modulator (218627_at), score: 0.51 DSCC1defective in sister chromatid cohesion 1 homolog (S. cerevisiae) (219000_s_at), score: -0.81 DSCR3Down syndrome critical region gene 3 (203635_at), score: 0.45 DSN1DSN1, MIND kinetochore complex component, homolog (S. cerevisiae) (219512_at), score: -0.88 DTLdenticleless homolog (Drosophila) (218585_s_at), score: -0.83 DUSP14dual specificity phosphatase 14 (203367_at), score: 0.41 DUSP5dual specificity phosphatase 5 (209457_at), score: 0.5 DVL3dishevelled, dsh homolog 3 (Drosophila) (201908_at), score: 0.48 E2F8E2F transcription factor 8 (219990_at), score: -0.78 EDN1endothelin 1 (218995_s_at), score: 0.57 EGR2early growth response 2 (Krox-20 homolog, Drosophila) (205249_at), score: 0.64 EGR3early growth response 3 (206115_at), score: 0.53 EIF2AK3eukaryotic translation initiation factor 2-alpha kinase 3 (218696_at), score: 0.4 EIF2C2eukaryotic translation initiation factor 2C, 2 (213310_at), score: 0.48 ENTPD7ectonucleoside triphosphate diphosphohydrolase 7 (220153_at), score: 0.73 ERAP1endoplasmic reticulum aminopeptidase 1 (214012_at), score: 0.4 ERCC6Lexcision repair cross-complementing rodent repair deficiency, complementation group 6-like (219650_at), score: -0.85 EREGepiregulin (205767_at), score: 0.45 EXO1exonuclease 1 (204603_at), score: -0.78 EZH2enhancer of zeste homolog 2 (Drosophila) (203358_s_at), score: -0.84 F10coagulation factor X (205620_at), score: 0.58 F3coagulation factor III (thromboplastin, tissue factor) (204363_at), score: 0.56 FABP3fatty acid binding protein 3, muscle and heart (mammary-derived growth inhibitor) (214285_at), score: 0.54 FADS3fatty acid desaturase 3 (216080_s_at), score: 0.4 FAM108B1family with sequence similarity 108, member B1 (220285_at), score: 0.49 FAM64Afamily with sequence similarity 64, member A (221591_s_at), score: -0.77 FANCAFanconi anemia, complementation group A (203805_s_at), score: -0.78 FANCGFanconi anemia, complementation group G (203564_at), score: -0.87 FANCIFanconi anemia, complementation group I (213007_at), score: -0.8 FASTKD5FAST kinase domains 5 (219016_at), score: 0.42 FBXO5F-box protein 5 (218875_s_at), score: -0.76 FBXW7F-box and WD repeat domain containing 7 (218751_s_at), score: 0.57 FCGRTFc fragment of IgG, receptor, transporter, alpha (218831_s_at), score: 0.43 FEM1Bfem-1 homolog b (C. elegans) (212367_at), score: 0.56 FGF1fibroblast growth factor 1 (acidic) (205117_at), score: 0.64 FGF2fibroblast growth factor 2 (basic) (204421_s_at), score: 0.42 FGF7fibroblast growth factor 7 (keratinocyte growth factor) (205782_at), score: 0.71 FOSBFBJ murine osteosarcoma viral oncogene homolog B (202768_at), score: 0.8 FOXM1forkhead box M1 (202580_x_at), score: -0.78 FOXO3Bforkhead box O3B pseudogene (210655_s_at), score: 0.5 FRYfurry homolog (Drosophila) (204072_s_at), score: 0.5 FUT4fucosyltransferase 4 (alpha (1,3) fucosyltransferase, myeloid-specific) (209892_at), score: 0.4 GABARAPL1GABA(A) receptor-associated protein like 1 (208868_s_at), score: 0.61 GABARAPL3GABA(A) receptors associated protein like 3 (pseudogene) (211458_s_at), score: 0.51 GADD45Agrowth arrest and DNA-damage-inducible, alpha (203725_at), score: 0.47 GALNAC4S-6STB cell RAG associated protein (203066_at), score: 0.49 GAP43growth associated protein 43 (204471_at), score: -0.77 GCC2GRIP and coiled-coil domain containing 2 (202832_at), score: 0.42 GCH1GTP cyclohydrolase 1 (204224_s_at), score: 0.62 GEMGTP binding protein overexpressed in skeletal muscle (204472_at), score: 0.78 GFPT2glutamine-fructose-6-phosphate transaminase 2 (205100_at), score: 0.55 GINS1GINS complex subunit 1 (Psf1 homolog) (206102_at), score: -0.9 GINS2GINS complex subunit 2 (Psf2 homolog) (221521_s_at), score: -0.9 GINS3GINS complex subunit 3 (Psf3 homolog) (45633_at), score: -0.86 GMNNgeminin, DNA replication inhibitor (218350_s_at), score: -0.83 GNPTABN-acetylglucosamine-1-phosphate transferase, alpha and beta subunits (212959_s_at), score: 0.59 GPNMBglycoprotein (transmembrane) nmb (201141_at), score: 0.61 GPR183G protein-coupled receptor 183 (205419_at), score: 0.82 GPR37G protein-coupled receptor 37 (endothelin receptor type B-like) (209631_s_at), score: 0.47 GRAMD3GRAM domain containing 3 (218706_s_at), score: 0.49 GSTM1glutathione S-transferase mu 1 (204550_x_at), score: 0.52 GTSE1G-2 and S-phase expressed 1 (204318_s_at), score: -0.81 HBEGFheparin-binding EGF-like growth factor (203821_at), score: 0.84 HIPK3homeodomain interacting protein kinase 3 (210148_at), score: 0.54 HIVEP1human immunodeficiency virus type I enhancer binding protein 1 (204512_at), score: 0.69 HIVEP2human immunodeficiency virus type I enhancer binding protein 2 (212642_s_at), score: 0.6 HK2hexokinase 2 (202934_at), score: 0.66 HLA-Cmajor histocompatibility complex, class I, C (211799_x_at), score: 0.45 HLA-Emajor histocompatibility complex, class I, E (200904_at), score: 0.42 HLA-Fmajor histocompatibility complex, class I, F (204806_x_at), score: 0.41 HLXH2.0-like homeobox (214438_at), score: 0.59 HMGB2high-mobility group box 2 (208808_s_at), score: -0.89 HMGB3L1high-mobility group box 3-like 1 (216548_x_at), score: -0.77 HMGCS13-hydroxy-3-methylglutaryl-Coenzyme A synthase 1 (soluble) (221750_at), score: 0.4 HMMRhyaluronan-mediated motility receptor (RHAMM) (207165_at), score: -0.81 HMOX1heme oxygenase (decycling) 1 (203665_at), score: 0.53 HOXA10homeobox A10 (213150_at), score: -0.84 HOXA9homeobox A9 (214651_s_at), score: -0.79 HSPA12Aheat shock 70kDa protein 12A (214434_at), score: 0.48 HSPB3heat shock 27kDa protein 3 (206375_s_at), score: 0.39 HTR2A5-hydroxytryptamine (serotonin) receptor 2A (207135_at), score: 0.62 ICAM1intercellular adhesion molecule 1 (202638_s_at), score: 0.82 IDI1isopentenyl-diphosphate delta isomerase 1 (204615_x_at), score: 0.65 IDI2isopentenyl-diphosphate delta isomerase 2 (217631_at), score: 0.49 IER3immediate early response 3 (201631_s_at), score: 0.41 IL11interleukin 11 (206924_at), score: 0.7 IL1Ainterleukin 1, alpha (210118_s_at), score: 0.4 IL1Binterleukin 1, beta (39402_at), score: 0.45 IL1RAPinterleukin 1 receptor accessory protein (205227_at), score: 0.57 IL1RL1interleukin 1 receptor-like 1 (207526_s_at), score: 0.49 IL6interleukin 6 (interferon, beta 2) (205207_at), score: 0.91 IL8interleukin 8 (202859_x_at), score: 0.61 INHBAinhibin, beta A (210511_s_at), score: 0.43 INSIG1insulin induced gene 1 (201627_s_at), score: 0.7 IRGQimmunity-related GTPase family, Q (64488_at), score: 0.45 ITGA1integrin, alpha 1 (214660_at), score: 0.63 JARID2jumonji, AT rich interactive domain 2 (203297_s_at), score: 0.69 JHDM1Djumonji C domain containing histone demethylase 1 homolog D (S. cerevisiae) (221778_at), score: 0.73 JMJD3jumonji domain containing 3, histone lysine demethylase (213146_at), score: 0.74 JMJD6jumonji domain containing 6 (212722_s_at), score: 0.65 JUNDjun D proto-oncogene (203751_x_at), score: 0.42 KIAA0247KIAA0247 (202181_at), score: 0.63 KIAA0586KIAA0586 (205631_at), score: -0.86 KIAA1024KIAA1024 (215081_at), score: 0.41 KIAA1128KIAA1128 (209379_s_at), score: 0.41 KIAA1644KIAA1644 (52837_at), score: 0.6 KIF11kinesin family member 11 (204444_at), score: -0.84 KIF15kinesin family member 15 (219306_at), score: -0.82 KIF18Bkinesin family member 18B (222039_at), score: -0.86 KIF20Bkinesin family member 20B (205235_s_at), score: -0.81 KIF22kinesin family member 22 (202183_s_at), score: -0.85 KIF2Ckinesin family member 2C (209408_at), score: -0.76 KIF4Akinesin family member 4A (218355_at), score: -0.83 KLHL18kelch-like 18 (Drosophila) (212882_at), score: 0.42 KLHL21kelch-like 21 (Drosophila) (203068_at), score: 0.59 LAMA5laminin, alpha 5 (210150_s_at), score: 0.43 LBHlimb bud and heart development homolog (mouse) (221011_s_at), score: 0.48 LDLRlow density lipoprotein receptor (202068_s_at), score: 0.43 LGALS3BPlectin, galactoside-binding, soluble, 3 binding protein (200923_at), score: 0.4 LHFPlipoma HMGIC fusion partner (218656_s_at), score: 0.51 LIFleukemia inhibitory factor (cholinergic differentiation factor) (205266_at), score: 0.9 LIG1ligase I, DNA, ATP-dependent (202726_at), score: -0.79 LIG4ligase IV, DNA, ATP-dependent (206235_at), score: 0.42 LIMS1LIM and senescent cell antigen-like domains 1 (207198_s_at), score: 0.43 LMBRD1LMBR1 domain containing 1 (218191_s_at), score: 0.59 LMCD1LIM and cysteine-rich domains 1 (218574_s_at), score: 0.8 LMNB1lamin B1 (203276_at), score: -0.84 LOH3CR2Aloss of heterozygosity, 3, chromosomal region 2, gene A (220244_at), score: 0.58 LPPR4plasticity related gene 1 (213496_at), score: 0.42 LRIG1leucine-rich repeats and immunoglobulin-like domains 1 (211596_s_at), score: 0.59 LRRC2leucine rich repeat containing 2 (219949_at), score: 0.4 LSM2LSM2 homolog, U6 small nuclear RNA associated (S. cerevisiae) (209449_at), score: -0.79 LUZP1leucine zipper protein 1 (221832_s_at), score: 0.53 LYSTlysosomal trafficking regulator (203518_at), score: 0.4 MAD2L1MAD2 mitotic arrest deficient-like 1 (yeast) (203362_s_at), score: -0.88 MAFv-maf musculoaponeurotic fibrosarcoma oncogene homolog (avian) (209348_s_at), score: 0.41 MAFFv-maf musculoaponeurotic fibrosarcoma oncogene homolog F (avian) (36711_at), score: 0.43 MAGI2membrane associated guanylate kinase, WW and PDZ domain containing 2 (209737_at), score: 0.53 MAN1C1mannosidase, alpha, class 1C, member 1 (218918_at), score: 0.61 MAP3K4mitogen-activated protein kinase kinase kinase 4 (216199_s_at), score: 0.55 MAP3K7IP1mitogen-activated protein kinase kinase kinase 7 interacting protein 1 (203901_at), score: -0.78 MARCH2membrane-associated ring finger (C3HC4) 2 (210075_at), score: 0.4 MAXMYC associated factor X (210734_x_at), score: 0.43 MCM10minichromosome maintenance complex component 10 (220651_s_at), score: -0.84 MCM2minichromosome maintenance complex component 2 (202107_s_at), score: -0.87 MCM3minichromosome maintenance complex component 3 (201555_at), score: -0.81 MCM4minichromosome maintenance complex component 4 (212141_at), score: -0.8 MCM5minichromosome maintenance complex component 5 (216237_s_at), score: -0.91 MCM6minichromosome maintenance complex component 6 (201930_at), score: -0.78 MCM7minichromosome maintenance complex component 7 (210983_s_at), score: -0.82 MDM1Mdm1 nuclear protein homolog (mouse) (213761_at), score: -0.79 MEF2Amyocyte enhancer factor 2A (214684_at), score: 0.44 MEIS1Meis homeobox 1 (204069_at), score: 0.42 MEIS3P1Meis homeobox 3 pseudogene 1 (214077_x_at), score: 0.51 MFAP3microfibrillar-associated protein 3 (213123_at), score: 0.45 MFAP4microfibrillar-associated protein 4 (212713_at), score: 0.6 MLXIPMLX interacting protein (202519_at), score: 0.43 MNS1meiosis-specific nuclear structural 1 (219703_at), score: -0.78 MSCmusculin (activated B-cell factor-1) (209928_s_at), score: 0.48 MSH2mutS homolog 2, colon cancer, nonpolyposis type 1 (E. coli) (209421_at), score: -0.79 MTSS1metastasis suppressor 1 (203037_s_at), score: 0.72 MYO1Emyosin IE (203072_at), score: 0.42 NAB1NGFI-A binding protein 1 (EGR1 binding protein 1) (208047_s_at), score: 0.44 NADSYN1NAD synthetase 1 (218840_s_at), score: 0.39 NAMPTnicotinamide phosphoribosyltransferase (217738_at), score: 0.62 NASPnuclear autoantigenic sperm protein (histone-binding) (201970_s_at), score: -0.76 NCAPD2non-SMC condensin I complex, subunit D2 (201774_s_at), score: -0.75 NCAPD3non-SMC condensin II complex, subunit D3 (212789_at), score: -0.88 NCAPGnon-SMC condensin I complex, subunit G (218663_at), score: -0.81 NCAPG2non-SMC condensin II complex, subunit G2 (219588_s_at), score: -0.76 NCAPHnon-SMC condensin I complex, subunit H (212949_at), score: -0.81 NCOA1nuclear receptor coactivator 1 (209106_at), score: 0.42 NDC80NDC80 homolog, kinetochore complex component (S. cerevisiae) (204162_at), score: -0.84 NDEL1nudE nuclear distribution gene E homolog (A. nidulans)-like 1 (208093_s_at), score: 0.52 NDRG1N-myc downstream regulated 1 (200632_s_at), score: 0.52 NEDD9neural precursor cell expressed, developmentally down-regulated 9 (202149_at), score: 0.61 NEIL3nei endonuclease VIII-like 3 (E. coli) (219502_at), score: -0.77 NEK2NIMA (never in mitosis gene a)-related kinase 2 (204641_at), score: -0.78 NFATC1nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 1 (210162_s_at), score: 0.86 NFIL3nuclear factor, interleukin 3 regulated (203574_at), score: 0.6 NFKB1nuclear factor of kappa light polypeptide gene enhancer in B-cells 1 (209239_at), score: 0.69 NFKB2nuclear factor of kappa light polypeptide gene enhancer in B-cells 2 (p49/p100) (207535_s_at), score: 0.44 NINJ1ninjurin 1 (203045_at), score: 0.56 NPnucleoside phosphorylase (201695_s_at), score: 0.41 NPC1Niemann-Pick disease, type C1 (202679_at), score: 0.51 NR3C1nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) (201866_s_at), score: 0.65 NR4A1nuclear receptor subfamily 4, group A, member 1 (202340_x_at), score: 0.71 NR4A2nuclear receptor subfamily 4, group A, member 2 (216248_s_at), score: 0.5 NR4A3nuclear receptor subfamily 4, group A, member 3 (209959_at), score: 0.89 NSMCE4Anon-SMC element 4 homolog A (S. cerevisiae) (219067_s_at), score: -0.75 NUPL1nucleoporin like 1 (204435_at), score: 0.5 NUSAP1nucleolar and spindle associated protein 1 (218039_at), score: -0.82 OIP5Opa interacting protein 5 (213599_at), score: -0.79 OLFML2Aolfactomedin-like 2A (213075_at), score: 0.54 ORC1Lorigin recognition complex, subunit 1-like (yeast) (205085_at), score: -0.8 ORC6Lorigin recognition complex, subunit 6 like (yeast) (219105_x_at), score: -0.86 OSMRoncostatin M receptor (205729_at), score: 0.47 P2RX4purinergic receptor P2X, ligand-gated ion channel, 4 (204088_at), score: 0.44 PANX1pannexin 1 (204715_at), score: 0.57 PARP2poly (ADP-ribose) polymerase 2 (214086_s_at), score: -0.82 PCNTpericentrin (203660_s_at), score: -0.77 PCTK2PCTAIRE protein kinase 2 (221918_at), score: 0.49 PDGFAplatelet-derived growth factor alpha polypeptide (205463_s_at), score: 0.73 PDGFDplatelet derived growth factor D (219304_s_at), score: 0.59 PDGFRBplatelet-derived growth factor receptor, beta polypeptide (202273_at), score: 0.46 PDLIM3PDZ and LIM domain 3 (209621_s_at), score: 0.49 PEG10paternally expressed 10 (212094_at), score: -0.76 PIGBphosphatidylinositol glycan anchor biosynthesis, class B (214151_s_at), score: 0.54 PIK3CDphosphoinositide-3-kinase, catalytic, delta polypeptide (203879_at), score: 0.72 PIONpigeon homolog (Drosophila) (222150_s_at), score: 0.46 PKMYT1protein kinase, membrane associated tyrosine/threonine 1 (204267_x_at), score: -0.82 PLAURplasminogen activator, urokinase receptor (211924_s_at), score: 0.4 PLCXD1phosphatidylinositol-specific phospholipase C, X domain containing 1 (218951_s_at), score: 0.43 PLEKHF2pleckstrin homology domain containing, family F (with FYVE domain) member 2 (218640_s_at), score: 0.41 PLK3polo-like kinase 3 (Drosophila) (204958_at), score: 0.55 PLSCR4phospholipid scramblase 4 (218901_at), score: 0.54 PMEPA1prostate transmembrane protein, androgen induced 1 (217875_s_at), score: 0.95 POLA1polymerase (DNA directed), alpha 1, catalytic subunit (204835_at), score: -0.88 POLA2polymerase (DNA directed), alpha 2 (70kD subunit) (204441_s_at), score: -0.83 POLD1polymerase (DNA directed), delta 1, catalytic subunit 125kDa (203422_at), score: -0.9 POLD3polymerase (DNA-directed), delta 3, accessory subunit (212836_at), score: -0.78 POLEpolymerase (DNA directed), epsilon (216026_s_at), score: -0.85 POLE2polymerase (DNA directed), epsilon 2 (p59 subunit) (205909_at), score: -0.82 POLQpolymerase (DNA directed), theta (219510_at), score: -0.78 POLR1Cpolymerase (RNA) I polypeptide C, 30kDa (207515_s_at), score: 0.49 PPARDperoxisome proliferator-activated receptor delta (37152_at), score: 0.62 PPLperiplakin (203407_at), score: 0.49 PPP1R15Aprotein phosphatase 1, regulatory (inhibitor) subunit 15A (202014_at), score: 0.4 PPP3CCprotein phosphatase 3 (formerly 2B), catalytic subunit, gamma isoform (207000_s_at), score: 0.4 PRIM1primase, DNA, polypeptide 1 (49kDa) (205053_at), score: -0.84 PRKG1protein kinase, cGMP-dependent, type I (207119_at), score: 0.47 PROS1protein S (alpha) (207808_s_at), score: 0.41 PRR11proline rich 11 (219392_x_at), score: -0.76 PSMC3IPPSMC3 interacting protein (213951_s_at), score: -0.83 PSRC1proline/serine-rich coiled-coil 1 (201896_s_at), score: -0.77 PTHLHparathyroid hormone-like hormone (211756_at), score: 0.62 PTPN1protein tyrosine phosphatase, non-receptor type 1 (202716_at), score: 0.41 PTTG1pituitary tumor-transforming 1 (203554_x_at), score: -0.76 RAB33ARAB33A, member RAS oncogene family (206039_at), score: 0.52 RABGEF1RAB guanine nucleotide exchange factor (GEF) 1 (218310_at), score: 0.41 RACGAP1Rac GTPase activating protein 1 (222077_s_at), score: -0.77 RAD51AP1RAD51 associated protein 1 (204146_at), score: -0.82 RAD54BRAD54 homolog B (S. cerevisiae) (219494_at), score: -0.82 RAD54LRAD54-like (S. cerevisiae) (204558_at), score: -0.82 RAD9ARAD9 homolog A (S. pombe) (204828_at), score: -0.8 RANBP2RAN binding protein 2 (201711_x_at), score: 0.4 RAPGEF2Rap guanine nucleotide exchange factor (GEF) 2 (203097_s_at), score: 0.55 RASGRP1RAS guanyl releasing protein 1 (calcium and DAG-regulated) (205590_at), score: 0.44 RCAN1regulator of calcineurin 1 (215253_s_at), score: 0.43 RCAN2regulator of calcineurin 2 (203498_at), score: 0.53 RCL1RNA terminal phosphate cyclase-like 1 (218544_s_at), score: 0.45 RELBv-rel reticuloendotheliosis viral oncogene homolog B (205205_at), score: 0.52 RFC3replication factor C (activator 1) 3, 38kDa (204127_at), score: -0.86 RFC4replication factor C (activator 1) 4, 37kDa (204023_at), score: -0.83 RGPD5RANBP2-like and GRIP domain containing 5 (210676_x_at), score: 0.58 RGS2regulator of G-protein signaling 2, 24kDa (202388_at), score: 0.67 RHOQras homolog gene family, member Q (212119_at), score: 0.43 RLFrearranged L-myc fusion (204243_at), score: 0.41 RMI1RMI1, RecQ mediated genome instability 1, homolog (S. cerevisiae) (218979_at), score: -0.79 RNASEH2Aribonuclease H2, subunit A (203022_at), score: -0.82 RNF103ring finger protein 103 (202636_at), score: 0.49 RNF25ring finger protein 25 (218861_at), score: 0.41 RPL39Lribosomal protein L39-like (210115_at), score: -0.78 RPRD2regulation of nuclear pre-mRNA domain containing 2 (212553_at), score: -0.82 RRADRas-related associated with diabetes (204803_s_at), score: 0.69 RUNX1runt-related transcription factor 1 (209360_s_at), score: 0.78 RWDD2ARWD domain containing 2A (213555_at), score: 0.4 SAMD4Asterile alpha motif domain containing 4A (212845_at), score: 0.42 SC4MOLsterol-C4-methyl oxidase-like (209146_at), score: 0.57 SC5DLsterol-C5-desaturase (ERG3 delta-5-desaturase homolog, S. cerevisiae)-like (211423_s_at), score: 0.4 SCARB1scavenger receptor class B, member 1 (201819_at), score: 0.42 SENP5SUMO1/sentrin specific peptidase 5 (213184_at), score: 0.43 SERPINB2serpin peptidase inhibitor, clade B (ovalbumin), member 2 (204614_at), score: 0.47 SERPINB8serpin peptidase inhibitor, clade B (ovalbumin), member 8 (206034_at), score: 0.42 SETXsenataxin (201964_at), score: 0.42 SGK1serum/glucocorticoid regulated kinase 1 (201739_at), score: 0.51 SH3TC2SH3 domain and tetratricopeptide repeats 2 (219710_at), score: -0.8 SHBSrc homology 2 domain containing adaptor protein B (204657_s_at), score: 0.44 SHCBP1SHC SH2-domain binding protein 1 (219493_at), score: -0.76 SKILSKI-like oncogene (206675_s_at), score: 0.72 SLC10A3solute carrier family 10 (sodium/bile acid cotransporter family), member 3 (204928_s_at), score: 0.39 SLC19A2solute carrier family 19 (thiamine transporter), member 2 (209681_at), score: 0.83 SLC20A2solute carrier family 20 (phosphate transporter), member 2 (202744_at), score: 0.4 SLC2A14solute carrier family 2 (facilitated glucose transporter), member 14 (222088_s_at), score: 0.54 SLC2A3P1solute carrier family 2 (facilitated glucose transporter), member 3 pseudogene 1 (221751_at), score: 0.51 SLC46A3solute carrier family 46, member 3 (214719_at), score: 0.41 SLC7A6solute carrier family 7 (cationic amino acid transporter, y+ system), member 6 (203578_s_at), score: 0.46 SMAD7SMAD family member 7 (204790_at), score: 0.46 SMC2structural maintenance of chromosomes 2 (204240_s_at), score: -0.77 SMOXspermine oxidase (210357_s_at), score: 0.76 SMURF1SMAD specific E3 ubiquitin protein ligase 1 (212666_at), score: 0.54 SOCS2suppressor of cytokine signaling 2 (203373_at), score: 0.42 SOX4SRY (sex determining region Y)-box 4 (201417_at), score: 0.64 SOX9SRY (sex determining region Y)-box 9 (202935_s_at), score: 0.65 SPATA2Lspermatogenesis associated 2-like (214965_at), score: 0.65 SPC25SPC25, NDC80 kinetochore complex component, homolog (S. cerevisiae) (209891_at), score: -0.8 SPHK1sphingosine kinase 1 (219257_s_at), score: 0.41 SPRED2sprouty-related, EVH1 domain containing 2 (212458_at), score: 0.66 SPRY2sprouty homolog 2 (Drosophila) (204011_at), score: 0.66 SPRY4sprouty homolog 4 (Drosophila) (221489_s_at), score: 0.63 SPSB1splA/ryanodine receptor domain and SOCS box containing 1 (219677_at), score: 0.56 SQLEsqualene epoxidase (213562_s_at), score: 0.72 SREBF2sterol regulatory element binding transcription factor 2 (201247_at), score: 0.51 SRFserum response factor (c-fos serum response element-binding transcription factor) (202401_s_at), score: 0.73 SS18L1synovial sarcoma translocation gene on chromosome 18-like 1 (213140_s_at), score: 0.44 ST3GAL1ST3 beta-galactoside alpha-2,3-sialyltransferase 1 (208322_s_at), score: 0.71 STBD1starch binding domain 1 (203986_at), score: 0.56 STILSCL/TAL1 interrupting locus (205339_at), score: -0.78 STK38Lserine/threonine kinase 38 like (212572_at), score: 0.91 STMN1stathmin 1/oncoprotein 18 (200783_s_at), score: -0.81 STX6syntaxin 6 (212799_at), score: 0.43 SUV39H1suppressor of variegation 3-9 homolog 1 (Drosophila) (218619_s_at), score: -0.76 SVEP1sushi, von Willebrand factor type A, EGF and pentraxin domain containing 1 (213247_at), score: 0.63 TACC3transforming, acidic coiled-coil containing protein 3 (218308_at), score: -0.79 TACSTD2tumor-associated calcium signal transducer 2 (202286_s_at), score: 0.47 TAPBPLTAP binding protein-like (218746_at), score: 0.47 TBX3T-box 3 (219682_s_at), score: 0.51 TGFBR1transforming growth factor, beta receptor 1 (206943_at), score: 0.76 THAP10THAP domain containing 10 (219596_at), score: -0.76 THBDthrombomodulin (203887_s_at), score: 0.7 TICAM1toll-like receptor adaptor molecule 1 (213191_at), score: 0.58 TIMELESStimeless homolog (Drosophila) (203046_s_at), score: -0.78 TK1thymidine kinase 1, soluble (202338_at), score: -0.85 TM2D1TM2 domain containing 1 (211703_s_at), score: 0.5 TM6SF1transmembrane 6 superfamily member 1 (219892_at), score: 0.63 TMEM194Atransmembrane protein 194A (212621_at), score: -0.8 TMEM2transmembrane protein 2 (218113_at), score: 0.42 TMEM39Atransmembrane protein 39A (218615_s_at), score: 0.6 TMEM41Btransmembrane protein 41B (212623_at), score: 0.67 TMEM87Atransmembrane protein 87A (212202_s_at), score: 0.5 TNFAIP2tumor necrosis factor, alpha-induced protein 2 (202510_s_at), score: 0.42 TNFAIP3tumor necrosis factor, alpha-induced protein 3 (202644_s_at), score: 0.55 TNFAIP6tumor necrosis factor, alpha-induced protein 6 (206026_s_at), score: 0.51 TNFAIP8tumor necrosis factor, alpha-induced protein 8 (210260_s_at), score: 0.47 TNFSF4tumor necrosis factor (ligand) superfamily, member 4 (207426_s_at), score: 0.58 TNS3tensin 3 (217853_at), score: 0.49 TNXBtenascin XB (216333_x_at), score: 0.41 TOM1target of myb1 (chicken) (202807_s_at), score: 0.49 TP53BP2tumor protein p53 binding protein, 2 (203120_at), score: 0.74 TPD52L1tumor protein D52-like 1 (203786_s_at), score: 0.49 TPP1tripeptidyl peptidase I (200742_s_at), score: 0.48 TPST1tyrosylprotein sulfotransferase 1 (204140_at), score: 0.48 TPX2TPX2, microtubule-associated, homolog (Xenopus laevis) (210052_s_at), score: -0.8 TRAF3TNF receptor-associated factor 3 (221571_at), score: 0.48 TRAPPC10trafficking protein particle complex 10 (209412_at), score: 0.5 TRIB1tribbles homolog 1 (Drosophila) (202241_at), score: 0.81 TRIM13tripartite motif-containing 13 (203659_s_at), score: 0.42 TRIM22tripartite motif-containing 22 (213293_s_at), score: 0.45 TRIM32tripartite motif-containing 32 (203846_at), score: 0.41 TRIP13thyroid hormone receptor interactor 13 (204033_at), score: -0.81 TRMT5TRM5 tRNA methyltransferase 5 homolog (S. cerevisiae) (221952_x_at), score: -0.79 TTC17tetratricopeptide repeat domain 17 (218972_at), score: 0.61 TTKTTK protein kinase (204822_at), score: -0.8 TWIST1twist homolog 1 (Drosophila) (213943_at), score: -0.8 UBA7ubiquitin-like modifier activating enzyme 7 (203281_s_at), score: 0.45 UNGuracil-DNA glycosylase (202330_s_at), score: -0.78 USP9Yubiquitin specific peptidase 9, Y-linked (fat facets-like, Drosophila) (206624_at), score: 0.41 USTuronyl-2-sulfotransferase (205139_s_at), score: 0.48 UTP3UTP3, small subunit (SSU) processome component, homolog (S. cerevisiae) (209486_at), score: 0.46 VCLvinculin (200930_s_at), score: 0.65 VDRvitamin D (1,25- dihydroxyvitamin D3) receptor (204255_s_at), score: 0.4 VEGFAvascular endothelial growth factor A (211527_x_at), score: 0.83 WDHD1WD repeat and HMG-box DNA binding protein 1 (216228_s_at), score: -0.82 WDR37WD repeat domain 37 (211383_s_at), score: 0.46 WDR76WD repeat domain 76 (205519_at), score: -0.77 WEE1WEE1 homolog (S. pombe) (212533_at), score: -0.9 WISP1WNT1 inducible signaling pathway protein 1 (206796_at), score: 0.45 WRAP53WD repeat containing, antisense to TP53 (44563_at), score: -0.81 WWTR1WW domain containing transcription regulator 1 (202132_at), score: 0.56 YPEL5yippee-like 5 (Drosophila) (217783_s_at), score: 0.46 YRDCyrdC domain containing (E. coli) (218647_s_at), score: 0.78 ZCCHC6zinc finger, CCHC domain containing 6 (220933_s_at), score: 0.4 ZFP36L2zinc finger protein 36, C3H type-like 2 (201367_s_at), score: -0.84 ZKSCAN3zinc finger with KRAB and SCAN domains 3 (211773_s_at), score: -0.78 ZNF143zinc finger protein 143 (221873_at), score: 0.5 ZNF184zinc finger protein 184 (213452_at), score: -0.79 ZNF263zinc finger protein 263 (203707_at), score: 0.41 ZNF304zinc finger protein 304 (207753_at), score: 0.4 ZNF35zinc finger protein 35 (206096_at), score: 0.65 ZNF672zinc finger protein 672 (218068_s_at), score: 0.69 ZNF85zinc finger protein 85 (206572_x_at), score: -0.77
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| 1Twin.CEL | 1 | 2 | DS-twin | hgu133plus2 | Down | DS-twin 1 |
| 5CTwin.CEL | 5 | 2 | DS-twin | hgu133plus2 | Down | DS-twin 5 |
| 6Twin.CEL | 6 | 2 | DS-twin | hgu133plus2 | none | DS-twin 6 |
| 2Twin.CEL | 2 | 2 | DS-twin | hgu133plus2 | none | DS-twin 2 |
| 46A.CEL | 1 | 3 | DS-mosaic | hgu133plus2 | none | DS-mosaic 1 |
| E-TABM-263-raw-cel-1515486331.cel | 35 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486351.cel | 36 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486231.cel | 30 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485851.cel | 11 | 6 | Cycle | hgu133a2 | none | Cycle 1 |