
Under-expression is coded with green,
over-expression with red color.

metabolic process
The chemical reactions and pathways, including anabolism and catabolism, by which living organisms transform chemical substances. Metabolic processes typically transform small molecules, but also include macromolecular processes such as DNA repair and replication, and protein synthesis and degradation.
organic acid metabolic process
The chemical reactions and pathways involving organic acids, any acidic compound containing carbon in covalent linkage.
lipid metabolic process
The chemical reactions and pathways involving lipids, compounds soluble in an organic solvent but not, or sparingly, in an aqueous solvent. Includes fatty acids; neutral fats, other fatty-acid esters, and soaps; long-chain (fatty) alcohols and waxes; sphingoids and other long-chain bases; glycolipids, phospholipids and sphingolipids; and carotenes, polyprenols, sterols, terpenes and other isoprenoids.
steroid catabolic process
The chemical reactions and pathways resulting in the breakdown of steroids, compounds with a 1,2,cyclopentanoperhydrophenanthrene nucleus.
biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
steroid metabolic process
The chemical reactions and pathways involving steroids, compounds with a 1,2,cyclopentanoperhydrophenanthrene nucleus.
bile acid metabolic process
The chemical reactions and pathways involving bile acids, any of a group of steroid carboxylic acids occurring in bile, where they are present as the sodium salts of their amides with glycine or taurine.
catabolic process
The chemical reactions and pathways resulting in the breakdown of substances, including the breakdown of carbon compounds with the liberation of energy for use by the cell or organism.
cellular process
Any process that is carried out at the cellular level, but not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
lipid catabolic process
The chemical reactions and pathways resulting in the breakdown of lipids, compounds soluble in an organic solvent but not, or sparingly, in an aqueous solvent.
organic acid catabolic process
The chemical reactions and pathways resulting in the breakdown of organic acids, any acidic compound containing carbon in covalent linkage.
carboxylic acid metabolic process
The chemical reactions and pathways involving carboxylic acids, any organic acid containing one or more carboxyl (COOH) groups or anions (COO-).
bile acid catabolic process
The chemical reactions and pathways resulting in the breakdown of bile acids, any of a group of steroid carboxylic acids occurring in bile.
monocarboxylic acid metabolic process
The chemical reactions and pathways involving monocarboxylic acids, any organic acid containing one carboxyl (COOH) group or anion (COO-).
cellular ketone metabolic process
The chemical reactions and pathways involving any of a class of organic compounds that contain the carbonyl group, CO, and in which the carbonyl group is bonded only to carbon atoms, as carried out by individual cells. The general formula for a ketone is RCOR, where R and R are alkyl or aryl groups.
oxoacid metabolic process
The chemical reactions and pathways involving any oxoacid; an oxoacid is a compound which contains oxygen, at least one other element, and at least one hydrogen bound to oxygen, and which produces a conjugate base by loss of positive hydrogen ion(s) (hydrons).
cellular metabolic process
The chemical reactions and pathways by which individual cells transform chemical substances.
primary metabolic process
The chemical reactions and pathways involving those compounds which are formed as a part of the normal anabolic and catabolic processes. These processes take place in most, if not all, cells of the organism.
cellular catabolic process
The chemical reactions and pathways resulting in the breakdown of substances, carried out by individual cells.
small molecule metabolic process
The chemical reactions and pathways involving small molecules, any monomeric molecule of small relative molecular mass.
small molecule catabolic process
The chemical reactions and pathways resulting in the breakdown of small molecules, any monomeric molecule of small relative molecular mass.
carboxylic acid catabolic process
The chemical reactions and pathways resulting in the breakdown of carboxylic acids, any organic acid containing one or more carboxyl (-COOH) groups.
all
NA
cellular metabolic process
The chemical reactions and pathways by which individual cells transform chemical substances.
cellular catabolic process
The chemical reactions and pathways resulting in the breakdown of substances, carried out by individual cells.
organic acid metabolic process
The chemical reactions and pathways involving organic acids, any acidic compound containing carbon in covalent linkage.
cellular ketone metabolic process
The chemical reactions and pathways involving any of a class of organic compounds that contain the carbonyl group, CO, and in which the carbonyl group is bonded only to carbon atoms, as carried out by individual cells. The general formula for a ketone is RCOR, where R and R are alkyl or aryl groups.
small molecule catabolic process
The chemical reactions and pathways resulting in the breakdown of small molecules, any monomeric molecule of small relative molecular mass.
organic acid catabolic process
The chemical reactions and pathways resulting in the breakdown of organic acids, any acidic compound containing carbon in covalent linkage.
organic acid catabolic process
The chemical reactions and pathways resulting in the breakdown of organic acids, any acidic compound containing carbon in covalent linkage.
oxoacid metabolic process
The chemical reactions and pathways involving any oxoacid; an oxoacid is a compound which contains oxygen, at least one other element, and at least one hydrogen bound to oxygen, and which produces a conjugate base by loss of positive hydrogen ion(s) (hydrons).
lipid catabolic process
The chemical reactions and pathways resulting in the breakdown of lipids, compounds soluble in an organic solvent but not, or sparingly, in an aqueous solvent.
steroid catabolic process
The chemical reactions and pathways resulting in the breakdown of steroids, compounds with a 1,2,cyclopentanoperhydrophenanthrene nucleus.
bile acid catabolic process
The chemical reactions and pathways resulting in the breakdown of bile acids, any of a group of steroid carboxylic acids occurring in bile.
carboxylic acid catabolic process
The chemical reactions and pathways resulting in the breakdown of carboxylic acids, any organic acid containing one or more carboxyl (-COOH) groups.
bile acid catabolic process
The chemical reactions and pathways resulting in the breakdown of bile acids, any of a group of steroid carboxylic acids occurring in bile.
bile acid metabolic process
The chemical reactions and pathways involving bile acids, any of a group of steroid carboxylic acids occurring in bile, where they are present as the sodium salts of their amides with glycine or taurine.

plasma membrane
The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins.
membrane
Double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
integral to membrane
Penetrating at least one phospholipid bilayer of a membrane. May also refer to the state of being buried in the bilayer with no exposure outside the bilayer. When used to describe a protein, indicates that all or part of the peptide sequence is embedded in the membrane.
extracellular region
The space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers the host cell environment outside an intracellular parasite.
integral to plasma membrane
Penetrating at least one phospholipid bilayer of a plasma membrane. May also refer to the state of being buried in the bilayer with no exposure outside the bilayer.
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
membrane attack complex
A protein complex produced by sequentially activated components of the complement cascade inserted into a target cell membrane and forming a pore leading to cell lysis via ion and water flow.
extracellular space
That part of a multicellular organism outside the cells proper, usually taken to be outside the plasma membranes, and occupied by fluid.
cell
The basic structural and functional unit of all organisms. Includes the plasma membrane and any external encapsulating structures such as the cell wall and cell envelope.
pore complex
Any small opening in a membrane that allows the passage of gases and/or liquids.
intrinsic to membrane
Located in a membrane such that some covalently attached portion of the gene product, for example part of a peptide sequence or some other covalently attached moiety such as a GPI anchor, spans or is embedded in one or both leaflets of the membrane.
intrinsic to plasma membrane
Located in the plasma membrane such that some covalently attached portion of the gene product, for example part of a peptide sequence or some other covalently attached moiety such as a GPI anchor, spans or is embedded in one or both leaflets of the membrane.
macromolecular complex
A stable assembly of two or more macromolecules, i.e. proteins, nucleic acids, carbohydrates or lipids, in which the constituent parts function together.
protein complex
Any macromolecular complex composed of two or more polypeptide subunits, which may or may not be identical. Protein complexes may have other associated non-protein prosthetic groups, such as nucleotides, metal ions or other small molecules.
extracellular region part
Any constituent part of the extracellular region, the space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers constituent parts of the host cell environment outside an intracellular parasite.
membrane part
Any constituent part of a membrane, a double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
plasma membrane part
Any constituent part of the plasma membrane, the membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins.
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
all
NA
extracellular region part
Any constituent part of the extracellular region, the space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers constituent parts of the host cell environment outside an intracellular parasite.
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
membrane part
Any constituent part of a membrane, a double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
plasma membrane part
Any constituent part of the plasma membrane, the membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins.
membrane attack complex
A protein complex produced by sequentially activated components of the complement cascade inserted into a target cell membrane and forming a pore leading to cell lysis via ion and water flow.
intrinsic to plasma membrane
Located in the plasma membrane such that some covalently attached portion of the gene product, for example part of a peptide sequence or some other covalently attached moiety such as a GPI anchor, spans or is embedded in one or both leaflets of the membrane.
pore complex
Any small opening in a membrane that allows the passage of gases and/or liquids.
integral to plasma membrane
Penetrating at least one phospholipid bilayer of a plasma membrane. May also refer to the state of being buried in the bilayer with no exposure outside the bilayer.
membrane attack complex
A protein complex produced by sequentially activated components of the complement cascade inserted into a target cell membrane and forming a pore leading to cell lysis via ion and water flow.

molecular_function
Elemental activities, such as catalysis or binding, describing the actions of a gene product at the molecular level. A given gene product may exhibit one or more molecular functions.
enzyme inhibitor activity
Stops, prevents or reduces the activity of an enzyme.
endopeptidase inhibitor activity
Stops, prevents or reduces the activity of an endopeptidase, any enzyme that hydrolyzes nonterminal peptide bonds in polypeptides.
endopeptidase regulator activity
Modulates the activity of a peptidase, any enzyme that hydrolyzes nonterminal peptide bonds in polypeptides.
peptidase inhibitor activity
Stops, prevents or reduces the activity of a peptidase, any enzyme that catalyzes the hydrolysis peptide bonds.
enzyme regulator activity
Modulates the activity of an enzyme.
peptidase regulator activity
Modulates the activity of a peptidase, any enzyme that catalyzes the hydrolysis peptide bonds.
all
NA
peptidase inhibitor activity
Stops, prevents or reduces the activity of a peptidase, any enzyme that catalyzes the hydrolysis peptide bonds.
endopeptidase inhibitor activity
Stops, prevents or reduces the activity of an endopeptidase, any enzyme that hydrolyzes nonterminal peptide bonds in polypeptides.
| Id | Pvalue | ExpCount | Count | Size | Term |
|---|---|---|---|---|---|
| 00830 | 1.064e-03 | 0.189 | 4 | 25 | Retinol metabolism |
| 04610 | 1.099e-02 | 0.378 | 4 | 50 | Complement and coagulation cascades |
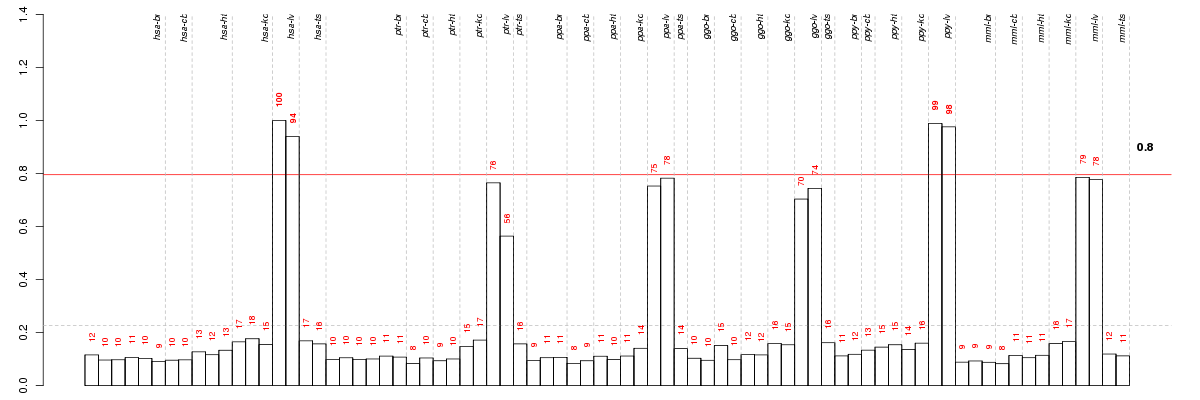
ADH4alcohol dehydrogenase 4 (class II), pi polypeptide (ENSG00000198099), score: 0.74 AKR1D1aldo-keto reductase family 1, member D1 (delta 4-3-ketosteroid-5-beta-reductase) (ENSG00000122787), score: 0.72 APCSamyloid P component, serum (ENSG00000132703), score: 0.71 APOFapolipoprotein F (ENSG00000175336), score: 0.79 BPIL1bactericidal/permeability-increasing protein-like 1 (ENSG00000078898), score: 0.83 C14orf21chromosome 14 open reading frame 21 (ENSG00000196943), score: 0.72 C17orf48chromosome 17 open reading frame 48 (ENSG00000170222), score: 0.73 C20orf194chromosome 20 open reading frame 194 (ENSG00000088854), score: -0.74 C20orf70chromosome 20 open reading frame 70 (ENSG00000131050), score: 0.74 C8Acomplement component 8, alpha polypeptide (ENSG00000157131), score: 0.73 C8Bcomplement component 8, beta polypeptide (ENSG00000021852), score: 0.72 CD7CD7 molecule (ENSG00000173762), score: 0.71 CHST4carbohydrate (N-acetylglucosamine 6-O) sulfotransferase 4 (ENSG00000140835), score: 0.78 CNGA1cyclic nucleotide gated channel alpha 1 (ENSG00000198515), score: 1 CPB2carboxypeptidase B2 (plasma) (ENSG00000080618), score: 0.72 CSTAcystatin A (stefin A) (ENSG00000121552), score: 0.82 CXCR6chemokine (C-X-C motif) receptor 6 (ENSG00000172215), score: 0.72 CYP2C8cytochrome P450, family 2, subfamily C, polypeptide 8 (ENSG00000138115), score: 0.74 DGAT1diacylglycerol O-acyltransferase 1 (ENSG00000185000), score: 0.7 EI24etoposide induced 2.4 mRNA (ENSG00000149547), score: 0.71 ENPP5ectonucleotide pyrophosphatase/phosphodiesterase 5 (putative) (ENSG00000112796), score: -0.73 ENPP7ectonucleotide pyrophosphatase/phosphodiesterase 7 (ENSG00000182156), score: 0.76 F9coagulation factor IX (ENSG00000101981), score: 0.72 FETUBfetuin B (ENSG00000090512), score: 0.73 FGF21fibroblast growth factor 21 (ENSG00000105550), score: 0.83 GJC3gap junction protein, gamma 3, 30.2kDa (ENSG00000176402), score: 0.74 GZMKgranzyme K (granzyme 3; tryptase II) (ENSG00000113088), score: 0.72 HALhistidine ammonia-lyase (ENSG00000084110), score: 0.79 HAO1hydroxyacid oxidase (glycolate oxidase) 1 (ENSG00000101323), score: 0.72 HSD17B13hydroxysteroid (17-beta) dehydrogenase 13 (ENSG00000170509), score: 0.72 IFNAR1interferon (alpha, beta and omega) receptor 1 (ENSG00000142166), score: 0.74 INHBCinhibin, beta C (ENSG00000175189), score: 0.73 INHBEinhibin, beta E (ENSG00000139269), score: 0.77 KRT27keratin 27 (ENSG00000171446), score: 0.87 LECT2leukocyte cell-derived chemotaxin 2 (ENSG00000145826), score: 0.74 LGALS4lectin, galactoside-binding, soluble, 4 (ENSG00000171747), score: 0.73 LONP2lon peptidase 2, peroxisomal (ENSG00000102910), score: 0.72 LRRC31leucine rich repeat containing 31 (ENSG00000114248), score: 0.95 LYPD2LY6/PLAUR domain containing 2 (ENSG00000197353), score: 0.75 MINPP1multiple inositol-polyphosphate phosphatase 1 (ENSG00000107789), score: 0.79 MLANAmelan-A (ENSG00000120215), score: 0.81 MMABmethylmalonic aciduria (cobalamin deficiency) cblB type (ENSG00000139428), score: 0.82 MSMBmicroseminoprotein, beta- (ENSG00000138294), score: 0.79 NCR1natural cytotoxicity triggering receptor 1 (ENSG00000189430), score: 0.73 ORM1orosomucoid 1 (ENSG00000229314), score: 0.72 OTCornithine carbamoyltransferase (ENSG00000036473), score: 0.72 PLEK2pleckstrin 2 (ENSG00000100558), score: 0.75 PRLprolactin (ENSG00000172179), score: 0.74 RDH16retinol dehydrogenase 16 (all-trans) (ENSG00000139547), score: 0.74 RFC2replication factor C (activator 1) 2, 40kDa (ENSG00000049541), score: 0.71 SCGB3A2secretoglobin, family 3A, member 2 (ENSG00000164265), score: 0.75 SERPINA10serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 10 (ENSG00000140093), score: 0.73 SERPINA11serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 11 (ENSG00000186910), score: 0.72 SERPINA4serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 4 (ENSG00000100665), score: 0.76 SERPINA7serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 7 (ENSG00000123561), score: 0.71 SLC10A1solute carrier family 10 (sodium/bile acid cotransporter family), member 1 (ENSG00000100652), score: 0.73 SLC13A5solute carrier family 13 (sodium-dependent citrate transporter), member 5 (ENSG00000141485), score: 0.8 SLC17A2solute carrier family 17 (sodium phosphate), member 2 (ENSG00000112337), score: 0.71 SLC25A47solute carrier family 25, member 47 (ENSG00000140107), score: 0.75 SPP2secreted phosphoprotein 2, 24kDa (ENSG00000072080), score: 0.81 SULT2A1sulfotransferase family, cytosolic, 2A, dehydroepiandrosterone (DHEA)-preferring, member 1 (ENSG00000105398), score: 0.72 TFF1trefoil factor 1 (ENSG00000160182), score: 0.78 TLX1T-cell leukemia homeobox 1 (ENSG00000107807), score: 0.72 TRAT1T cell receptor associated transmembrane adaptor 1 (ENSG00000163519), score: 0.75 TRPM8transient receptor potential cation channel, subfamily M, member 8 (ENSG00000144481), score: 0.82 TTPALtocopherol (alpha) transfer protein-like (ENSG00000124120), score: 0.82 UROC1urocanase domain containing 1 (ENSG00000159650), score: 0.71 USH2AUsher syndrome 2A (autosomal recessive, mild) (ENSG00000042781), score: 0.83 ZG16zymogen granule protein 16 homolog (rat) (ENSG00000174992), score: 0.83
| Id | species | tissue | sex | individual |
|---|---|---|---|---|
| hsa_lv_m2_ca1 | hsa | lv | m | 2 |
| ppy_lv_f_ca1 | ppy | lv | f | _ |
| ppy_lv_m_ca1 | ppy | lv | m | _ |
| hsa_lv_m1_ca1 | hsa | lv | m | 1 |