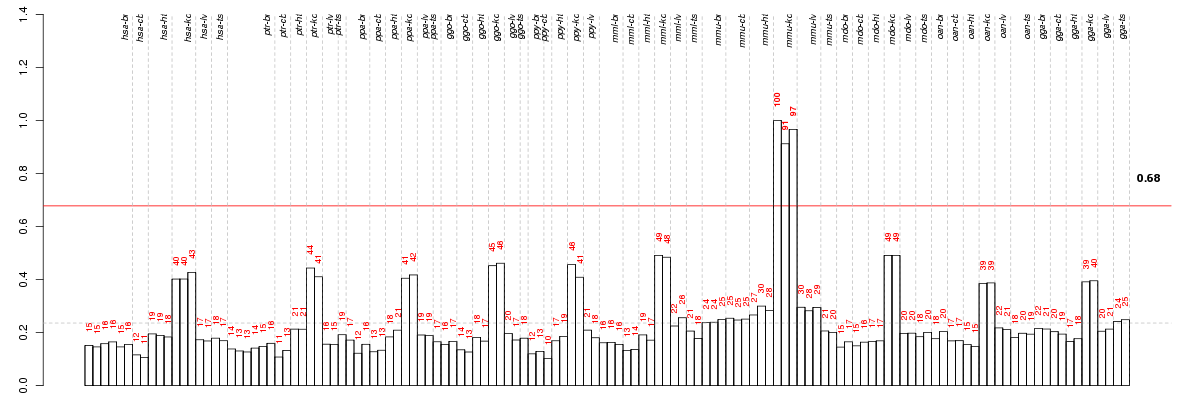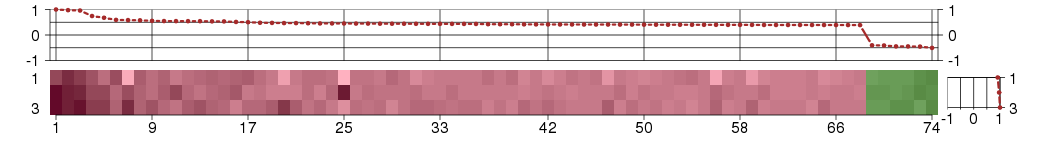

Under-expression is coded with green,
over-expression with red color.


extracellular region
The space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers the host cell environment outside an intracellular parasite.
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
extracellular space
That part of a multicellular organism outside the cells proper, usually taken to be outside the plasma membranes, and occupied by fluid.
extracellular matrix
A structure lying external to one or more cells, which provides structural support for cells or tissues; may be completely external to the cell (as in animals) or be part of the cell (as in plants).
extracellular matrix part
Any constituent part of the extracellular matrix, the structure lying external to one or more cells, which provides structural support for cells or tissues; may be completely external to the cell (as in animals) or be part of the cell (as often seen in plants).
extracellular region part
Any constituent part of the extracellular region, the space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers constituent parts of the host cell environment outside an intracellular parasite.
all
NA
extracellular region part
Any constituent part of the extracellular region, the space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers constituent parts of the host cell environment outside an intracellular parasite.
extracellular matrix part
Any constituent part of the extracellular matrix, the structure lying external to one or more cells, which provides structural support for cells or tissues; may be completely external to the cell (as in animals) or be part of the cell (as often seen in plants).

| Id | Pvalue | ExpCount | Count | Size | Term |
|---|---|---|---|---|---|
| 00601 | 3.429e-03 | 0.08022 | 3 | 7 | Glycosphingolipid biosynthesis - lacto and neolacto series |
A4GNTalpha-1,4-N-acetylglucosaminyltransferase (ENSG00000118017), score: 1 ANKRD13Cankyrin repeat domain 13C (ENSG00000118454), score: 0.48 AOAHacyloxyacyl hydrolase (neutrophil) (ENSG00000136250), score: 0.54 ARSBarylsulfatase B (ENSG00000113273), score: 0.45 ATP11AATPase, class VI, type 11A (ENSG00000068650), score: 0.47 B3GALT5UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 5 (ENSG00000183778), score: 0.44 B4GALT2UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 2 (ENSG00000117411), score: -0.45 BNC2basonuclin 2 (ENSG00000173068), score: 0.4 BPNT13'(2'), 5'-bisphosphate nucleotidase 1 (ENSG00000162813), score: 0.58 C11orf54chromosome 11 open reading frame 54 (ENSG00000182919), score: 0.39 CASRcalcium-sensing receptor (ENSG00000036828), score: 0.39 CCRL1chemokine (C-C motif) receptor-like 1 (ENSG00000129048), score: 0.39 CDCP1CUB domain containing protein 1 (ENSG00000163814), score: 0.44 COL10A1collagen, type X, alpha 1 (ENSG00000123500), score: 0.54 COL4A3collagen, type IV, alpha 3 (Goodpasture antigen) (ENSG00000169031), score: 0.41 COL4A4collagen, type IV, alpha 4 (ENSG00000081052), score: 0.45 DKK2dickkopf homolog 2 (Xenopus laevis) (ENSG00000155011), score: 0.41 EGFepidermal growth factor (ENSG00000138798), score: 0.53 EGFL6EGF-like-domain, multiple 6 (ENSG00000198759), score: 0.51 ENTPD4ectonucleoside triphosphate diphosphohydrolase 4 (ENSG00000197217), score: 0.39 ERBB2IPerbb2 interacting protein (ENSG00000112851), score: 0.39 FBXO31F-box protein 31 (ENSG00000103264), score: -0.4 FGFBP1fibroblast growth factor binding protein 1 (ENSG00000137440), score: 0.42 FUCA2fucosidase, alpha-L- 2, plasma (ENSG00000001036), score: 0.44 FUT9fucosyltransferase 9 (alpha (1,3) fucosyltransferase) (ENSG00000172461), score: 0.4 GAS2growth arrest-specific 2 (ENSG00000148935), score: 0.43 GBA2glucosidase, beta (bile acid) 2 (ENSG00000070610), score: -0.45 GCNT1glucosaminyl (N-acetyl) transferase 1, core 2 (ENSG00000187210), score: 0.68 H1FXH1 histone family, member X (ENSG00000184897), score: -0.51 HOXA10homeobox A10 (ENSG00000153807), score: 0.4 HOXB7homeobox B7 (ENSG00000120087), score: 0.39 IL5RAinterleukin 5 receptor, alpha (ENSG00000091181), score: 0.47 ILDR1immunoglobulin-like domain containing receptor 1 (ENSG00000145103), score: 0.41 ITGB6integrin, beta 6 (ENSG00000115221), score: 0.41 KCNJ1potassium inwardly-rectifying channel, subfamily J, member 1 (ENSG00000151704), score: 0.4 KLklotho (ENSG00000133116), score: 0.44 KRT80keratin 80 (ENSG00000167767), score: 0.59 LRRC31leucine rich repeat containing 31 (ENSG00000114248), score: 0.44 MED28mediator complex subunit 28 (ENSG00000118579), score: 0.42 MEP1Ameprin A, alpha (PABA peptide hydrolase) (ENSG00000112818), score: 0.96 MEP1Bmeprin A, beta (ENSG00000141434), score: 0.97 MMP13matrix metallopeptidase 13 (collagenase 3) (ENSG00000137745), score: 0.58 MYO6myosin VI (ENSG00000196586), score: 0.41 NOX4NADPH oxidase 4 (ENSG00000086991), score: 0.45 NSL1NSL1, MIND kinetochore complex component, homolog (S. cerevisiae) (ENSG00000117697), score: -0.41 NUDT19nudix (nucleoside diphosphate linked moiety X)-type motif 19 (ENSG00000213965), score: 0.54 OLFM4olfactomedin 4 (ENSG00000102837), score: 0.5 OSBPL3oxysterol binding protein-like 3 (ENSG00000070882), score: 0.39 PKD2polycystic kidney disease 2 (autosomal dominant) (ENSG00000118762), score: 0.4 PLAUplasminogen activator, urokinase (ENSG00000122861), score: 0.45 PPP6R2protein phosphatase 6, regulatory subunit 2 (ENSG00000100239), score: -0.45 RRHretinal pigment epithelium-derived rhodopsin homolog (ENSG00000180245), score: 0.45 SIM2single-minded homolog 2 (Drosophila) (ENSG00000159263), score: 0.42 SLC10A2solute carrier family 10 (sodium/bile acid cotransporter family), member 2 (ENSG00000125255), score: 0.46 SLC13A1solute carrier family 13 (sodium/sulfate symporters), member 1 (ENSG00000081800), score: 0.44 SLC18A1solute carrier family 18 (vesicular monoamine), member 1 (ENSG00000036565), score: 0.74 SLC22A4solute carrier family 22 (organic cation/ergothioneine transporter), member 4 (ENSG00000197208), score: 0.4 SLC25A17solute carrier family 25 (mitochondrial carrier; peroxisomal membrane protein, 34kDa), member 17 (ENSG00000100372), score: 0.39 SLC34A1solute carrier family 34 (sodium phosphate), member 1 (ENSG00000131183), score: 0.39 SLC9A8solute carrier family 9 (sodium/hydrogen exchanger), member 8 (ENSG00000197818), score: 0.4 SNX29sorting nexin 29 (ENSG00000048471), score: 0.41 SNX6sorting nexin 6 (ENSG00000129515), score: 0.41 SOSTDC1sclerostin domain containing 1 (ENSG00000171243), score: 0.44 TMBIM4transmembrane BAX inhibitor motif containing 4 (ENSG00000155957), score: 0.41 TMEM27transmembrane protein 27 (ENSG00000147003), score: 0.42 TMEM64transmembrane protein 64 (ENSG00000180694), score: 0.4 TMIGD1transmembrane and immunoglobulin domain containing 1 (ENSG00000182271), score: 0.55 TNFAIP8tumor necrosis factor, alpha-induced protein 8 (ENSG00000145779), score: 0.55 UPK3Auroplakin 3A (ENSG00000100373), score: 0.53 WDR89WD repeat domain 89 (ENSG00000140006), score: 0.4 ZBTB41zinc finger and BTB domain containing 41 (ENSG00000177888), score: 0.44 ZC3H12Czinc finger CCCH-type containing 12C (ENSG00000149289), score: 0.42 ZNF750zinc finger protein 750 (ENSG00000141579), score: 0.4 ZPLD1zona pellucida-like domain containing 1 (ENSG00000170044), score: 0.47
| Id | species | tissue | sex | individual |
|---|---|---|---|---|
| mmu_kd_m2_ca1 | mmu | kd | m | 2 |
| mmu_kd_f_ca1 | mmu | kd | f | _ |
| mmu_kd_m1_ca1 | mmu | kd | m | 1 |