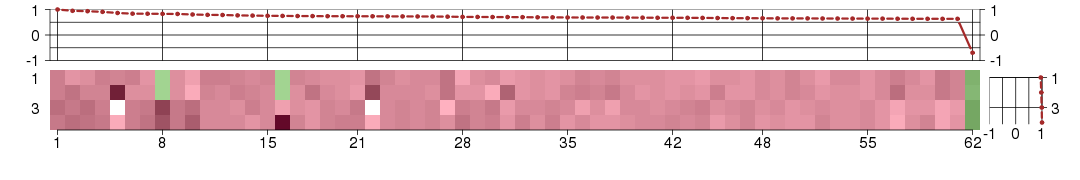

Under-expression is coded with green,
over-expression with red color.

metabolic process
The chemical reactions and pathways, including anabolism and catabolism, by which living organisms transform chemical substances. Metabolic processes typically transform small molecules, but also include macromolecular processes such as DNA repair and replication, and protein synthesis and degradation.
generation of precursor metabolites and energy
The chemical reactions and pathways resulting in the formation of precursor metabolites, substances from which energy is derived, and any process involved in the liberation of energy from these substances.
oxidation reduction
The process of removal or addition of one or more electrons with or without the concomitant removal or addition of a proton or protons.
electron transport chain
A process whereby a series of electron carriers operate together to transfer electrons from donors to any of several different terminal electron acceptors to generate a transmembrane electrochemical gradient.
biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
cellular process
Any process that is carried out at the cellular level, but not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
energy derivation by oxidation of organic compounds
The chemical reactions and pathways by which a cell derives energy from organic compounds; results in the oxidation of the compounds from which energy is released.
cellular metabolic process
The chemical reactions and pathways by which individual cells transform chemical substances.
cellular respiration
The enzymatic release of energy from organic compounds (especially carbohydrates and fats) which either requires oxygen (aerobic respiration) or does not (anaerobic respiration).
all
NA
cellular metabolic process
The chemical reactions and pathways by which individual cells transform chemical substances.
electron transport chain
A process whereby a series of electron carriers operate together to transfer electrons from donors to any of several different terminal electron acceptors to generate a transmembrane electrochemical gradient.

intracellular
The living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
contractile fiber
Fibers, composed of actin, myosin, and associated proteins, found in cells of smooth or striated muscle.
membrane
Double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
mitochondrion
A semiautonomous, self replicating organelle that occurs in varying numbers, shapes, and sizes in the cytoplasm of virtually all eukaryotic cells. It is notably the site of tissue respiration.
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
cell
The basic structural and functional unit of all organisms. Includes the plasma membrane and any external encapsulating structures such as the cell wall and cell envelope.
cytoplasm
All of the contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures.
mitochondrial envelope
The double lipid bilayer enclosing the mitochondrion and separating its contents from the cell cytoplasm; includes the intermembrane space.
mitochondrial inner membrane
The inner, i.e. lumen-facing, lipid bilayer of the mitochondrial envelope. It is highly folded to form cristae.
mitochondrial inner membrane presequence translocase complex
The protein transport machinery of the mitochondrial inner membrane that contains three essential Tim proteins: Tim17 and Tim23 are thought to build a preprotein translocation channel while Tim44 interacts transiently with the matrix heat-shock protein Hsp70 to form an ATP-driven import motor.
mitochondrial respiratory chain
The protein complexes that form the mitochondrial electron transport system (the respiratory chain), associated with the inner mitochondrial membrane. The respiratory chain complexes transfer electrons from an electron donor to an electron acceptor and are associated with a proton pump to create a transmembrane electrochemical gradient.
mitochondrial respiratory chain complex I
A protein complex located in the mitochondrial inner membrane that forms part of the mitochondrial respiratory chain. It contains about 25 different polypeptide subunits, including NADH dehydrogenase (ubiquinone), flavin mononucleotide and several different iron-sulfur clusters containing non-heme iron. The iron undergoes oxidation-reduction between Fe(II) and Fe(III), and catalyzes proton translocation linked to the oxidation of NADH by ubiquinone.
organelle inner membrane
The inner, i.e. lumen-facing, lipid bilayer of an organelle envelope; usually highly selective to most ions and metabolites.
myofibril
The contractile element of skeletal and cardiac muscle; a long, highly organized bundle of actin, myosin, and other proteins that contracts by a sliding filament mechanism.
sarcomere
The repeating unit of a myofibril in a muscle cell, composed of an array of overlapping thick and thin filaments between two adjacent Z discs.
NADH dehydrogenase complex
An integral membrane complex that possesses NADH oxidoreductase activity. The complex is one of the components of the electron transport chain. It catalyzes the transfer of a pair of electrons from NADH to a quinone.
organelle membrane
The lipid bilayer surrounding an organelle.
mitochondrial membrane
Either of the lipid bilayers that surround the mitochondrion and form the mitochondrial envelope.
organelle envelope
A double membrane structure enclosing an organelle, including two lipid bilayers and the region between them. In some cases, an organelle envelope may have more than two membranes.
envelope
A multilayered structure surrounding all or part of a cell; encompasses one or more lipid bilayers, and may include a cell wall layer; also includes the space between layers.
macromolecular complex
A stable assembly of two or more macromolecules, i.e. proteins, nucleic acids, carbohydrates or lipids, in which the constituent parts function together.
organelle
Organized structure of distinctive morphology and function. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton, and prokaryotic structures such as anammoxosomes and pirellulosomes. Excludes the plasma membrane.
membrane-bounded organelle
Organized structure of distinctive morphology and function, bounded by a single or double lipid bilayer membrane. Includes the nucleus, mitochondria, plastids, vacuoles, and vesicles. Excludes the plasma membrane.
non-membrane-bounded organelle
Organized structure of distinctive morphology and function, not bounded by a lipid bilayer membrane. Includes ribosomes, the cytoskeleton and chromosomes.
intracellular organelle
Organized structure of distinctive morphology and function, occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton. Excludes the plasma membrane.
intracellular membrane-bounded organelle
Organized structure of distinctive morphology and function, bounded by a single or double lipid bilayer membrane and occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, and vesicles. Excludes the plasma membrane.
intracellular non-membrane-bounded organelle
Organized structure of distinctive morphology and function, not bounded by a lipid bilayer membrane and occurring within the cell. Includes ribosomes, the cytoskeleton and chromosomes.
protein complex
Any macromolecular complex composed of two or more polypeptide subunits, which may or may not be identical. Protein complexes may have other associated non-protein prosthetic groups, such as nucleotides, metal ions or other small molecules.
organelle part
Any constituent part of an organelle, an organized structure of distinctive morphology and function. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton, but excludes the plasma membrane.
intracellular part
Any constituent part of the living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
membrane part
Any constituent part of a membrane, a double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
mitochondrial part
Any constituent part of a mitochondrion, a semiautonomous, self replicating organelle that occurs in varying numbers, shapes, and sizes in the cytoplasm of virtually all eukaryotic cells. It is notably the site of tissue respiration.
cytoplasmic part
Any constituent part of the cytoplasm, all of the contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures.
intracellular organelle part
A constituent part of an intracellular organelle, an organized structure of distinctive morphology and function, occurring within the cell. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton but excludes the plasma membrane.
contractile fiber part
Any constituent part of a contractile fiber, a fiber composed of actin, myosin, and associated proteins, found in cells of smooth or striated muscle.
mitochondrial membrane part
Any constituent part of a mitochondrial membrane, either of the lipid bilayers that surround the mitochondrion and form the mitochondrial envelope.
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
respiratory chain complex I
Respiratory chain complex I is an enzyme of the respiratory chain. It consists of at least 34 polypeptide chains and is L-shaped, with a horizontal arm lying in the membrane and a vertical arm that projects into the matrix. The electrons of NADH enter the chain at this complex.
respiratory chain
The protein complexes that form the electron transport system (the respiratory chain), associated with a cell membrane, usually the plasma membrane (in prokaryotes) or the inner mitochondrial membrane (on eukaryotes). The respiratory chain complexes transfer electrons from an electron donor to an electron acceptor and are associated with a proton pump to create a transmembrane electrochemical gradient.
all
NA
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
organelle part
Any constituent part of an organelle, an organized structure of distinctive morphology and function. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton, but excludes the plasma membrane.
organelle membrane
The lipid bilayer surrounding an organelle.
intracellular membrane-bounded organelle
Organized structure of distinctive morphology and function, bounded by a single or double lipid bilayer membrane and occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, and vesicles. Excludes the plasma membrane.
intracellular non-membrane-bounded organelle
Organized structure of distinctive morphology and function, not bounded by a lipid bilayer membrane and occurring within the cell. Includes ribosomes, the cytoskeleton and chromosomes.
intracellular organelle part
A constituent part of an intracellular organelle, an organized structure of distinctive morphology and function, occurring within the cell. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton but excludes the plasma membrane.
organelle envelope
A double membrane structure enclosing an organelle, including two lipid bilayers and the region between them. In some cases, an organelle envelope may have more than two membranes.
intracellular part
Any constituent part of the living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
organelle membrane
The lipid bilayer surrounding an organelle.
membrane part
Any constituent part of a membrane, a double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
organelle envelope
A double membrane structure enclosing an organelle, including two lipid bilayers and the region between them. In some cases, an organelle envelope may have more than two membranes.
organelle inner membrane
The inner, i.e. lumen-facing, lipid bilayer of an organelle envelope; usually highly selective to most ions and metabolites.
NADH dehydrogenase complex
An integral membrane complex that possesses NADH oxidoreductase activity. The complex is one of the components of the electron transport chain. It catalyzes the transfer of a pair of electrons from NADH to a quinone.
intracellular organelle
Organized structure of distinctive morphology and function, occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton. Excludes the plasma membrane.
intracellular organelle part
A constituent part of an intracellular organelle, an organized structure of distinctive morphology and function, occurring within the cell. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton but excludes the plasma membrane.
NADH dehydrogenase complex
An integral membrane complex that possesses NADH oxidoreductase activity. The complex is one of the components of the electron transport chain. It catalyzes the transfer of a pair of electrons from NADH to a quinone.
organelle inner membrane
The inner, i.e. lumen-facing, lipid bilayer of an organelle envelope; usually highly selective to most ions and metabolites.
mitochondrial inner membrane
The inner, i.e. lumen-facing, lipid bilayer of the mitochondrial envelope. It is highly folded to form cristae.
mitochondrial membrane part
Any constituent part of a mitochondrial membrane, either of the lipid bilayers that surround the mitochondrion and form the mitochondrial envelope.
mitochondrial envelope
The double lipid bilayer enclosing the mitochondrion and separating its contents from the cell cytoplasm; includes the intermembrane space.
mitochondrial membrane
Either of the lipid bilayers that surround the mitochondrion and form the mitochondrial envelope.
mitochondrial membrane part
Any constituent part of a mitochondrial membrane, either of the lipid bilayers that surround the mitochondrion and form the mitochondrial envelope.
cytoplasmic part
Any constituent part of the cytoplasm, all of the contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures.
contractile fiber
Fibers, composed of actin, myosin, and associated proteins, found in cells of smooth or striated muscle.
mitochondrion
A semiautonomous, self replicating organelle that occurs in varying numbers, shapes, and sizes in the cytoplasm of virtually all eukaryotic cells. It is notably the site of tissue respiration.
mitochondrial part
Any constituent part of a mitochondrion, a semiautonomous, self replicating organelle that occurs in varying numbers, shapes, and sizes in the cytoplasm of virtually all eukaryotic cells. It is notably the site of tissue respiration.
contractile fiber part
Any constituent part of a contractile fiber, a fiber composed of actin, myosin, and associated proteins, found in cells of smooth or striated muscle.
mitochondrial inner membrane presequence translocase complex
The protein transport machinery of the mitochondrial inner membrane that contains three essential Tim proteins: Tim17 and Tim23 are thought to build a preprotein translocation channel while Tim44 interacts transiently with the matrix heat-shock protein Hsp70 to form an ATP-driven import motor.
mitochondrial respiratory chain
The protein complexes that form the mitochondrial electron transport system (the respiratory chain), associated with the inner mitochondrial membrane. The respiratory chain complexes transfer electrons from an electron donor to an electron acceptor and are associated with a proton pump to create a transmembrane electrochemical gradient.
respiratory chain complex I
Respiratory chain complex I is an enzyme of the respiratory chain. It consists of at least 34 polypeptide chains and is L-shaped, with a horizontal arm lying in the membrane and a vertical arm that projects into the matrix. The electrons of NADH enter the chain at this complex.
mitochondrial respiratory chain complex I
A protein complex located in the mitochondrial inner membrane that forms part of the mitochondrial respiratory chain. It contains about 25 different polypeptide subunits, including NADH dehydrogenase (ubiquinone), flavin mononucleotide and several different iron-sulfur clusters containing non-heme iron. The iron undergoes oxidation-reduction between Fe(II) and Fe(III), and catalyzes proton translocation linked to the oxidation of NADH by ubiquinone.
mitochondrial membrane
Either of the lipid bilayers that surround the mitochondrion and form the mitochondrial envelope.
mitochondrial part
Any constituent part of a mitochondrion, a semiautonomous, self replicating organelle that occurs in varying numbers, shapes, and sizes in the cytoplasm of virtually all eukaryotic cells. It is notably the site of tissue respiration.
contractile fiber part
Any constituent part of a contractile fiber, a fiber composed of actin, myosin, and associated proteins, found in cells of smooth or striated muscle.
mitochondrial inner membrane presequence translocase complex
The protein transport machinery of the mitochondrial inner membrane that contains three essential Tim proteins: Tim17 and Tim23 are thought to build a preprotein translocation channel while Tim44 interacts transiently with the matrix heat-shock protein Hsp70 to form an ATP-driven import motor.
mitochondrial respiratory chain
The protein complexes that form the mitochondrial electron transport system (the respiratory chain), associated with the inner mitochondrial membrane. The respiratory chain complexes transfer electrons from an electron donor to an electron acceptor and are associated with a proton pump to create a transmembrane electrochemical gradient.
mitochondrial respiratory chain complex I
A protein complex located in the mitochondrial inner membrane that forms part of the mitochondrial respiratory chain. It contains about 25 different polypeptide subunits, including NADH dehydrogenase (ubiquinone), flavin mononucleotide and several different iron-sulfur clusters containing non-heme iron. The iron undergoes oxidation-reduction between Fe(II) and Fe(III), and catalyzes proton translocation linked to the oxidation of NADH by ubiquinone.
sarcomere
The repeating unit of a myofibril in a muscle cell, composed of an array of overlapping thick and thin filaments between two adjacent Z discs.

molecular_function
Elemental activities, such as catalysis or binding, describing the actions of a gene product at the molecular level. A given gene product may exhibit one or more molecular functions.
catalytic activity
Catalysis of a biochemical reaction at physiological temperatures. In biologically catalyzed reactions, the reactants are known as substrates, and the catalysts are naturally occurring macromolecular substances known as enzymes. Enzymes possess specific binding sites for substrates, and are usually composed wholly or largely of protein, but RNA that has catalytic activity (ribozyme) is often also regarded as enzymatic.
NADH dehydrogenase activity
Catalysis of the reaction: NADH + H+ + acceptor = NAD+ + reduced acceptor.
cytidine deaminase activity
Catalysis of the reaction: cytidine + H2O = uridine + NH3.
oxidoreductase activity
Catalysis of an oxidation-reduction (redox) reaction, a reversible chemical reaction in which the oxidation state of an atom or atoms within a molecule is altered. One substrate acts as a hydrogen or electron donor and becomes oxidized, while the other acts as hydrogen or electron acceptor and becomes reduced.
NADH dehydrogenase (ubiquinone) activity
Catalysis of the reaction: NADH + H+ + ubiquinone = NAD+ + ubiquinol.
oxidoreductase activity, acting on NADH or NADPH
Catalysis of an oxidation-reduction (redox) reaction in which NADH or NADPH acts as a hydrogen or electron donor and reduces a hydrogen or electron acceptor.
oxidoreductase activity, acting on NADH or NADPH, quinone or similar compound as acceptor
Catalysis of an oxidation-reduction (redox) reaction in which NADH or NADPH acts as a hydrogen or electron donor and reduces a quinone or a similar acceptor molecule.
hydrolase activity
Catalysis of the hydrolysis of various bonds, e.g. C-O, C-N, C-C, phosphoric anhydride bonds, etc. Hydrolase is the systematic name for any enzyme of EC class 3.
hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds
Catalysis of the hydrolysis of any carbon-nitrogen bond, C-N, with the exception of peptide bonds.
hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amidines
Catalysis of the hydrolysis of any non-peptide carbon-nitrogen bond in a cyclic amidine, a compound of the form R-C(=NH)-NH2.
deaminase activity
Catalysis of the removal of an amino group from a substrate, producing ammonia (NH3).
NADH dehydrogenase (quinone) activity
Catalysis of the reaction: NADH + H+ + a quinone = NAD+ + a quinol.
all
NA
NADH dehydrogenase (quinone) activity
Catalysis of the reaction: NADH + H+ + a quinone = NAD+ + a quinol.
cytidine deaminase activity
Catalysis of the reaction: cytidine + H2O = uridine + NH3.
| Id | Pvalue | ExpCount | Count | Size | Term |
|---|---|---|---|---|---|
| 05010 | 1.875e-02 | 0.6634 | 5 | 44 | Alzheimer's disease |
| 04260 | 1.996e-02 | 0.377 | 4 | 25 | Cardiac muscle contraction |
| 05012 | 2.485e-02 | 0.4071 | 4 | 27 | Parkinson's disease |
| 00190 | 3.317e-02 | 0.4524 | 4 | 30 | Oxidative phosphorylation |
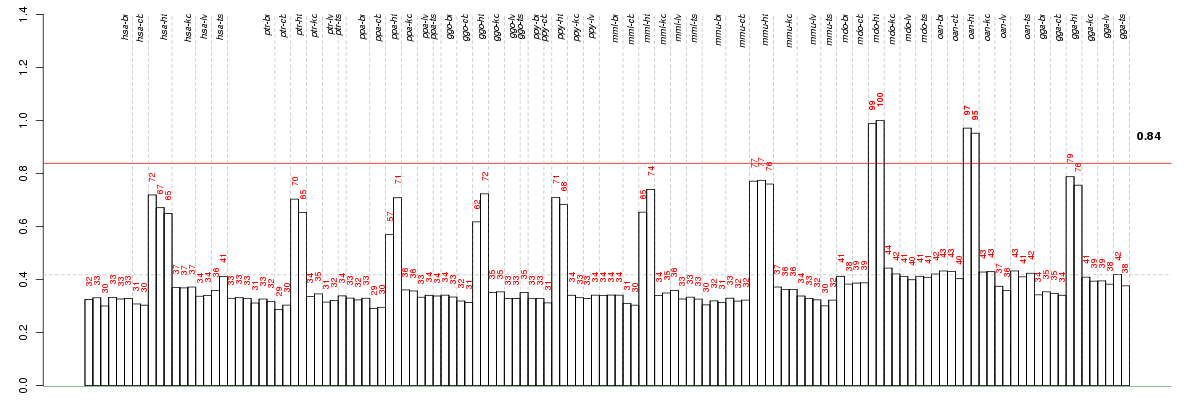
ABRAactin-binding Rho activating protein (ENSG00000174429), score: 0.64 ACAD8acyl-CoA dehydrogenase family, member 8 (ENSG00000151498), score: 0.71 ACANaggrecan (ENSG00000157766), score: 0.83 AICDAactivation-induced cytidine deaminase (ENSG00000111732), score: 0.75 AMPD1adenosine monophosphate deaminase 1 (ENSG00000116748), score: 0.86 AP3S1adaptor-related protein complex 3, sigma 1 subunit (ENSG00000177879), score: -0.7 APOBEC2apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 2 (ENSG00000124701), score: 0.64 AQRaquarius homolog (mouse) (ENSG00000021776), score: 0.78 ATG16L1ATG16 autophagy related 16-like 1 (S. cerevisiae) (ENSG00000085978), score: 0.71 ATG4AATG4 autophagy related 4 homolog A (S. cerevisiae) (ENSG00000101844), score: 0.69 ATP1B4ATPase, Na+/K+ transporting, beta 4 polypeptide (ENSG00000101892), score: 0.83 ATP2A2ATPase, Ca++ transporting, cardiac muscle, slow twitch 2 (ENSG00000174437), score: 0.66 C12orf5chromosome 12 open reading frame 5 (ENSG00000078237), score: 0.65 C6orf57chromosome 6 open reading frame 57 (ENSG00000154079), score: 0.67 CMYA5cardiomyopathy associated 5 (ENSG00000164309), score: 0.64 DUSP27dual specificity phosphatase 27 (putative) (ENSG00000198842), score: 0.65 EDNRAendothelin receptor type A (ENSG00000151617), score: 0.67 FBP2fructose-1,6-bisphosphatase 2 (ENSG00000130957), score: 0.74 FBXO40F-box protein 40 (ENSG00000163833), score: 0.66 FGF4fibroblast growth factor 4 (ENSG00000075388), score: 0.8 FGF6fibroblast growth factor 6 (ENSG00000111241), score: 0.73 FHL3four and a half LIM domains 3 (ENSG00000183386), score: 0.84 FSD2fibronectin type III and SPRY domain containing 2 (ENSG00000186628), score: 0.74 GATA5GATA binding protein 5 (ENSG00000130700), score: 0.83 GCNT7glucosaminyl (N-acetyl) transferase family member 7 (ENSG00000124091), score: 1 GJA3gap junction protein, alpha 3, 46kDa (ENSG00000121743), score: 0.73 GRXCR1glutaredoxin, cysteine rich 1 (ENSG00000215203), score: 0.7 GTPBP8GTP-binding protein 8 (putative) (ENSG00000163607), score: 0.74 GYG1glycogenin 1 (ENSG00000163754), score: 0.65 KCNJ2potassium inwardly-rectifying channel, subfamily J, member 2 (ENSG00000123700), score: 0.77 KLHL31kelch-like 31 (Drosophila) (ENSG00000124743), score: 0.68 LRRC10leucine rich repeat containing 10 (ENSG00000198812), score: 0.91 MYBPC3myosin binding protein C, cardiac (ENSG00000134571), score: 0.68 MYLK3myosin light chain kinase 3 (ENSG00000140795), score: 0.73 MYOTmyotilin (ENSG00000120729), score: 0.93 NDUFA10NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 10, 42kDa (ENSG00000130414), score: 0.65 NDUFA12NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 12 (ENSG00000184752), score: 0.68 NDUFA6NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 6, 14kDa (ENSG00000184983), score: 0.7 NDUFS3NADH dehydrogenase (ubiquinone) Fe-S protein 3, 30kDa (NADH-coenzyme Q reductase) (ENSG00000213619), score: 0.79 OXSR1oxidative-stress responsive 1 (ENSG00000172939), score: 0.64 PDHBpyruvate dehydrogenase (lipoamide) beta (ENSG00000168291), score: 0.69 PHEXphosphate regulating endopeptidase homolog, X-linked (ENSG00000102174), score: 0.76 PHF5APHD finger protein 5A (ENSG00000100410), score: 0.74 PPIP5K2diphosphoinositol pentakisphosphate kinase 2 (ENSG00000145725), score: 0.7 PPP1R3Dprotein phosphatase 1, regulatory (inhibitor) subunit 3D (ENSG00000132825), score: 0.74 RTN4IP1reticulon 4 interacting protein 1 (ENSG00000130347), score: 0.75 SLC25A3solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 3 (ENSG00000075415), score: 0.73 SMPXsmall muscle protein, X-linked (ENSG00000091482), score: 0.68 SUPV3L1suppressor of var1, 3-like 1 (S. cerevisiae) (ENSG00000156502), score: 0.69 TBX20T-box 20 (ENSG00000164532), score: 0.73 TBX5T-box 5 (ENSG00000089225), score: 0.63 TECRLtrans-2,3-enoyl-CoA reductase-like (ENSG00000205678), score: 0.64 TECTBtectorin beta (ENSG00000119913), score: 0.95 TIMM10translocase of inner mitochondrial membrane 10 homolog (yeast) (ENSG00000134809), score: 0.63 TIMM17Atranslocase of inner mitochondrial membrane 17 homolog A (yeast) (ENSG00000134375), score: 0.67 TNNT2troponin T type 2 (cardiac) (ENSG00000118194), score: 0.68 TRIM55tripartite motif-containing 55 (ENSG00000147573), score: 0.65 TRIM63tripartite motif-containing 63 (ENSG00000158022), score: 0.64 TSG101tumor susceptibility gene 101 (ENSG00000074319), score: 0.68 TXLNBtaxilin beta (ENSG00000164440), score: 0.66 UQCRC2ubiquinol-cytochrome c reductase core protein II (ENSG00000140740), score: 0.69 WNT11wingless-type MMTV integration site family, member 11 (ENSG00000085741), score: 0.72
| Id | species | tissue | sex | individual |
|---|---|---|---|---|
| oan_ht_f_ca1 | oan | ht | f | _ |
| oan_ht_m_ca1 | oan | ht | m | _ |
| mdo_ht_m_ca1 | mdo | ht | m | _ |
| mdo_ht_f_ca1 | mdo | ht | f | _ |