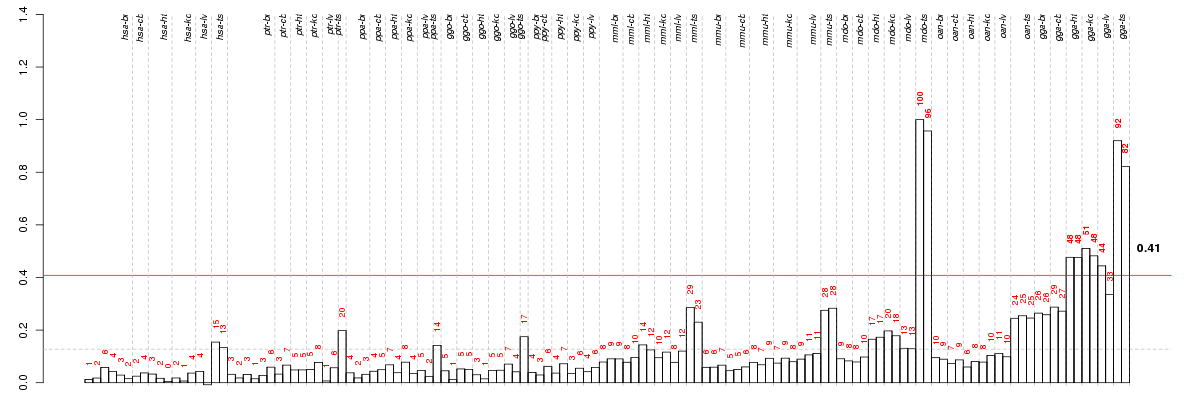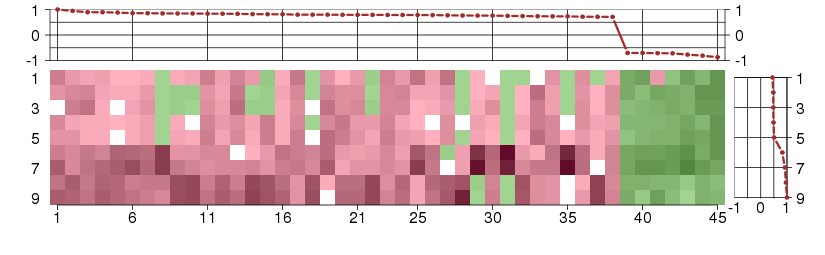

Under-expression is coded with green,
over-expression with red color.



ACER1alkaline ceramidase 1 (ENSG00000167769), score: 0.84 ADSLadenylosuccinate lyase (ENSG00000100354), score: 0.78 AIREautoimmune regulator (ENSG00000160224), score: 0.82 APBB3amyloid beta (A4) precursor protein-binding, family B, member 3 (ENSG00000113108), score: -0.87 APTXaprataxin (ENSG00000137074), score: 0.8 ARHGAP40Rho GTPase activating protein 40 (ENSG00000124143), score: 0.71 BRD4bromodomain containing 4 (ENSG00000141867), score: 0.72 C20orf79chromosome 20 open reading frame 79 (ENSG00000132631), score: 0.84 CIB3calcium and integrin binding family member 3 (ENSG00000141977), score: 0.79 CMAScytidine monophosphate N-acetylneuraminic acid synthetase (ENSG00000111726), score: -0.81 CRYBA1crystallin, beta A1 (ENSG00000108255), score: 0.76 CXorf22chromosome X open reading frame 22 (ENSG00000165164), score: 0.85 E2F8E2F transcription factor 8 (ENSG00000129173), score: 0.88 ENPP4ectonucleotide pyrophosphatase/phosphodiesterase 4 (putative) (ENSG00000001561), score: 0.85 ERGIC3ERGIC and golgi 3 (ENSG00000125991), score: -0.72 ERI1exoribonuclease 1 (ENSG00000104626), score: 0.73 FKBP14FK506 binding protein 14, 22 kDa (ENSG00000106080), score: 0.83 FOXN1forkhead box N1 (ENSG00000109101), score: 0.76 GALNT5UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 5 (GalNAc-T5) (ENSG00000136542), score: 0.74 GORASP1golgi reassembly stacking protein 1, 65kDa (ENSG00000114745), score: -0.71 GPR1G protein-coupled receptor 1 (ENSG00000183671), score: 0.94 HEATR6HEAT repeat containing 6 (ENSG00000068097), score: 0.81 IMPG2interphotoreceptor matrix proteoglycan 2 (ENSG00000081148), score: 0.8 KIAA1609KIAA1609 (ENSG00000140950), score: 0.71 KIF18Bkinesin family member 18B (ENSG00000186185), score: 0.86 MPLmyeloproliferative leukemia virus oncogene (ENSG00000117400), score: 0.83 MYBv-myb myeloblastosis viral oncogene homolog (avian) (ENSG00000118513), score: 0.84 MYF5myogenic factor 5 (ENSG00000111049), score: 0.75 NUP54nucleoporin 54kDa (ENSG00000138750), score: 0.78 PDIK1LPDLIM1 interacting kinase 1 like (ENSG00000175087), score: 0.79 PLRG1pleiotropic regulator 1 (PRL1 homolog, Arabidopsis) (ENSG00000171566), score: -0.77 POLE2polymerase (DNA directed), epsilon 2 (p59 subunit) (ENSG00000100479), score: 0.8 PUS3pseudouridylate synthase 3 (ENSG00000110060), score: 0.89 RAB3GAP1RAB3 GTPase activating protein subunit 1 (catalytic) (ENSG00000115839), score: -0.71 RFX6regulatory factor X, 6 (ENSG00000185002), score: 0.73 RPE65retinal pigment epithelium-specific protein 65kDa (ENSG00000116745), score: 0.74 SEMA3Dsema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3D (ENSG00000153993), score: 0.79 TERTtelomerase reverse transcriptase (ENSG00000164362), score: 0.79 TMEM26transmembrane protein 26 (ENSG00000196932), score: 0.89 TP63tumor protein p63 (ENSG00000073282), score: 0.78 TRAF3IP3TRAF3 interacting protein 3 (ENSG00000009790), score: 0.77 UFSP2UFM1-specific peptidase 2 (ENSG00000109775), score: -0.7 UHRF1ubiquitin-like with PHD and ring finger domains 1 (ENSG00000034063), score: 0.81 VASH2vasohibin 2 (ENSG00000143494), score: 0.76
| Id | species | tissue | sex | individual |
|---|---|---|---|---|
| gga_lv_m_ca1 | gga | lv | m | _ |
| gga_ht_f_ca1 | gga | ht | f | _ |
| gga_ht_m_ca1 | gga | ht | m | _ |
| gga_kd_f_ca1 | gga | kd | f | _ |
| gga_kd_m_ca1 | gga | kd | m | _ |
| gga_ts_m2_ca1 | gga | ts | m | 2 |
| gga_ts_m1_ca1 | gga | ts | m | 1 |
| mdo_ts_m1_ca1 | mdo | ts | m | 1 |
| mdo_ts_m2_ca1 | mdo | ts | m | 2 |