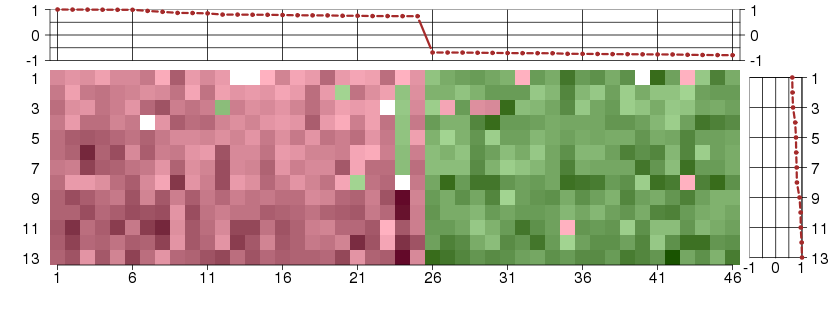

Under-expression is coded with green,
over-expression with red color.



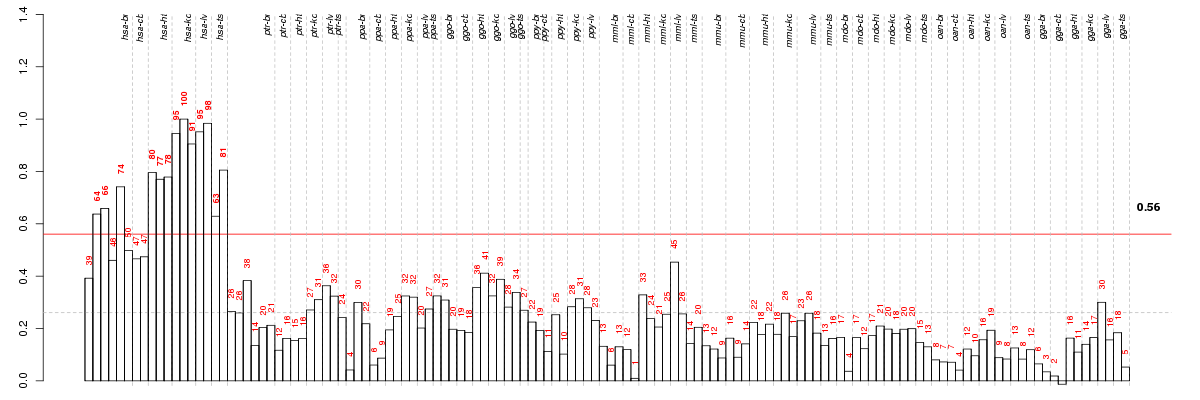
AIMP2aminoacyl tRNA synthetase complex-interacting multifunctional protein 2 (ENSG00000106305), score: 0.74 ALS2CR4amyotrophic lateral sclerosis 2 (juvenile) chromosome region, candidate 4 (ENSG00000155755), score: -0.71 APPBP2amyloid beta precursor protein (cytoplasmic tail) binding protein 2 (ENSG00000062725), score: -0.76 ATG10ATG10 autophagy related 10 homolog (S. cerevisiae) (ENSG00000152348), score: 0.95 ATG4CATG4 autophagy related 4 homolog C (S. cerevisiae) (ENSG00000125703), score: -0.7 C1orf107chromosome 1 open reading frame 107 (ENSG00000117597), score: -0.76 C5orf41chromosome 5 open reading frame 41 (ENSG00000164463), score: -0.78 CCNT2cyclin T2 (ENSG00000082258), score: -0.76 CHCHD8coiled-coil-helix-coiled-coil-helix domain containing 8 (ENSG00000181924), score: 0.77 CIITAclass II, major histocompatibility complex, transactivator (ENSG00000179583), score: 0.99 CISD2CDGSH iron sulfur domain 2 (ENSG00000145354), score: 0.75 COMMD2COMM domain containing 2 (ENSG00000114744), score: -0.79 DLEC1deleted in lung and esophageal cancer 1 (ENSG00000008226), score: 0.78 DPH1DPH1 homolog (S. cerevisiae) (ENSG00000108963), score: 0.85 EMILIN3elastin microfibril interfacer 3 (ENSG00000183798), score: 0.79 EXOSC7exosome component 7 (ENSG00000075914), score: 0.8 GPCPD1glycerophosphocholine phosphodiesterase GDE1 homolog (S. cerevisiae) (ENSG00000125772), score: -0.69 GPR114G protein-coupled receptor 114 (ENSG00000159618), score: 0.74 HECTD2HECT domain containing 2 (ENSG00000165338), score: -0.72 HEMK1HemK methyltransferase family member 1 (ENSG00000114735), score: 0.77 HMBOX1homeobox containing 1 (ENSG00000147421), score: -0.71 HOOK3hook homolog 3 (Drosophila) (ENSG00000168172), score: -0.74 INTS2integrator complex subunit 2 (ENSG00000108506), score: -0.69 LIN7Clin-7 homolog C (C. elegans) (ENSG00000148943), score: -0.74 MAP4K3mitogen-activated protein kinase kinase kinase kinase 3 (ENSG00000011566), score: -0.71 METTL6methyltransferase like 6 (ENSG00000206562), score: -0.78 MMABmethylmalonic aciduria (cobalamin deficiency) cblB type (ENSG00000139428), score: 0.8 NAA25N(alpha)-acetyltransferase 25, NatB auxiliary subunit (ENSG00000111300), score: -0.7 PABPC1Lpoly(A) binding protein, cytoplasmic 1-like (ENSG00000101104), score: 0.86 PLEKHJ1pleckstrin homology domain containing, family J member 1 (ENSG00000104886), score: 0.99 QARSglutaminyl-tRNA synthetase (ENSG00000172053), score: 0.75 RSPO1R-spondin homolog (Xenopus laevis) (ENSG00000169218), score: 1 SCAMP1secretory carrier membrane protein 1 (ENSG00000085365), score: -0.74 SLC18A2solute carrier family 18 (vesicular monoamine), member 2 (ENSG00000165646), score: 0.91 SLC19A1solute carrier family 19 (folate transporter), member 1 (ENSG00000173638), score: 0.79 SNX13sorting nexin 13 (ENSG00000071189), score: -0.79 SNX22sorting nexin 22 (ENSG00000157734), score: 1 SUN1Sad1 and UNC84 domain containing 1 (ENSG00000164828), score: 0.74 TBC1D8TBC1 domain family, member 8 (with GRAM domain) (ENSG00000204634), score: 0.99 TH1LTH1-like (Drosophila) (ENSG00000101158), score: 1 TRDMT1tRNA aspartic acid methyltransferase 1 (ENSG00000107614), score: -0.75 TTC15tetratricopeptide repeat domain 15 (ENSG00000171853), score: 0.87 WBP1WW domain binding protein 1 (ENSG00000115274), score: 0.77 WDR1WD repeat domain 1 (ENSG00000071127), score: 0.74 ZBTB6zinc finger and BTB domain containing 6 (ENSG00000186130), score: -0.75 ZNF292zinc finger protein 292 (ENSG00000188994), score: -0.69
| Id | species | tissue | sex | individual |
|---|---|---|---|---|
| hsa_ts_m2_ca1 | hsa | ts | m | 2 |
| hsa_br_m3_ca1 | hsa | br | m | 3 |
| hsa_br_m6_ca1 | hsa | br | m | 6 |
| hsa_br_m7_ca1 | hsa | br | m | 7 |
| hsa_ht_m2_ca1 | hsa | ht | m | 2 |
| hsa_ht_f_ca1 | hsa | ht | f | _ |
| hsa_ht_m1_ca1 | hsa | ht | m | 1 |
| hsa_ts_m1_ca1 | hsa | ts | m | 1 |
| hsa_kd_f_ca1 | hsa | kd | f | _ |
| hsa_kd_m2_ca1 | hsa | kd | m | 2 |
| hsa_lv_m1_ca1 | hsa | lv | m | 1 |
| hsa_lv_m2_ca1 | hsa | lv | m | 2 |
| hsa_kd_m1_ca1 | hsa | kd | m | 1 |