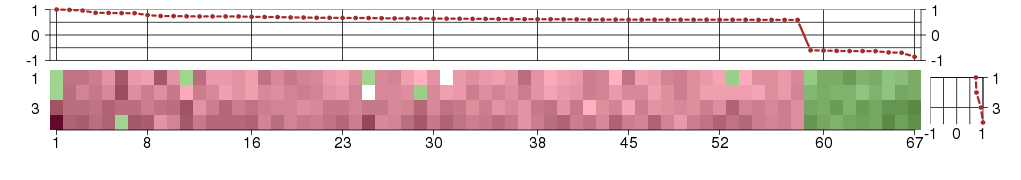

Under-expression is coded with green,
over-expression with red color.



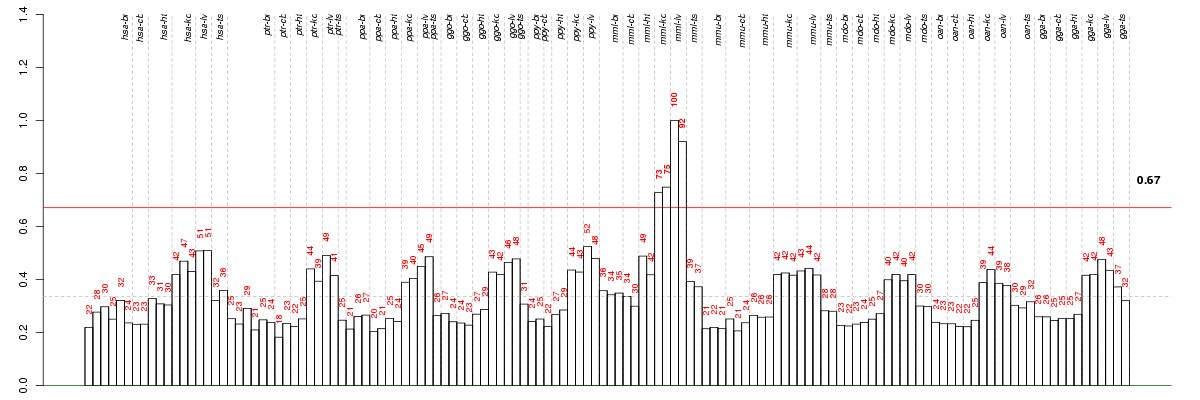
ACER1alkaline ceramidase 1 (ENSG00000167769), score: 0.62 ACSF3acyl-CoA synthetase family member 3 (ENSG00000176715), score: 0.6 AGXTalanine-glyoxylate aminotransferase (ENSG00000172482), score: 0.59 ALG2asparagine-linked glycosylation 2, alpha-1,3-mannosyltransferase homolog (S. cerevisiae) (ENSG00000119523), score: 0.73 ANXA10annexin A10 (ENSG00000109511), score: 0.96 AQP8aquaporin 8 (ENSG00000103375), score: 0.64 BCAR3breast cancer anti-estrogen resistance 3 (ENSG00000137936), score: 0.69 C20orf7chromosome 20 open reading frame 7 (ENSG00000101247), score: -0.86 C2orf54chromosome 2 open reading frame 54 (ENSG00000172478), score: 0.67 CASP9caspase 9, apoptosis-related cysteine peptidase (ENSG00000132906), score: 0.63 CCDC28Bcoiled-coil domain containing 28B (ENSG00000160050), score: -0.63 CDC42BPACDC42 binding protein kinase alpha (DMPK-like) (ENSG00000143776), score: -0.68 CLDN14claudin 14 (ENSG00000159261), score: 0.85 CLINT1clathrin interactor 1 (ENSG00000113282), score: 0.71 CYFIP1cytoplasmic FMR1 interacting protein 1 (ENSG00000068793), score: 0.73 D2HGDHD-2-hydroxyglutarate dehydrogenase (ENSG00000180902), score: 0.63 DKK2dickkopf homolog 2 (Xenopus laevis) (ENSG00000155011), score: 0.65 DLK1delta-like 1 homolog (Drosophila) (ENSG00000185559), score: 0.63 DNAJC7DnaJ (Hsp40) homolog, subfamily C, member 7 (ENSG00000168259), score: -0.7 EDEM1ER degradation enhancer, mannosidase alpha-like 1 (ENSG00000134109), score: 0.62 EME1essential meiotic endonuclease 1 homolog 1 (S. pombe) (ENSG00000154920), score: 0.6 EPHX1epoxide hydrolase 1, microsomal (xenobiotic) (ENSG00000143819), score: 0.6 ERLIN2ER lipid raft associated 2 (ENSG00000147475), score: 0.59 ESPNLespin-like (ENSG00000144488), score: 0.78 ESR1estrogen receptor 1 (ENSG00000091831), score: 0.59 F2RL2coagulation factor II (thrombin) receptor-like 2 (ENSG00000164220), score: 0.71 FPGSfolylpolyglutamate synthase (ENSG00000136877), score: 0.64 FZD5frizzled homolog 5 (Drosophila) (ENSG00000163251), score: 0.61 HEMK1HemK methyltransferase family member 1 (ENSG00000114735), score: 0.6 IGSF6immunoglobulin superfamily, member 6 (ENSG00000140749), score: 0.74 IL1R2interleukin 1 receptor, type II (ENSG00000115590), score: 0.73 IVDisovaleryl-CoA dehydrogenase (ENSG00000128928), score: 0.62 KBTBD2kelch repeat and BTB (POZ) domain containing 2 (ENSG00000170852), score: -0.63 KIAA1841KIAA1841 (ENSG00000162929), score: -0.61 KLHL14kelch-like 14 (Drosophila) (ENSG00000197705), score: 0.73 LGMNlegumain (ENSG00000100600), score: 0.68 MCM10minichromosome maintenance complex component 10 (ENSG00000065328), score: 0.74 MOBKL2BMOB1, Mps One Binder kinase activator-like 2B (yeast) (ENSG00000120162), score: 0.62 MON1AMON1 homolog A (yeast) (ENSG00000164077), score: 0.62 NSDHLNAD(P) dependent steroid dehydrogenase-like (ENSG00000147383), score: 0.67 NT5M5',3'-nucleotidase, mitochondrial (ENSG00000205309), score: -0.63 OXNAD1oxidoreductase NAD-binding domain containing 1 (ENSG00000154814), score: 0.65 PLCG2phospholipase C, gamma 2 (phosphatidylinositol-specific) (ENSG00000197943), score: 0.6 POLR3Epolymerase (RNA) III (DNA directed) polypeptide E (80kD) (ENSG00000058600), score: 0.6 PPAPDC1Bphosphatidic acid phosphatase type 2 domain containing 1B (ENSG00000147535), score: 0.87 PTBP2polypyrimidine tract binding protein 2 (ENSG00000117569), score: -0.6 RPP40ribonuclease P/MRP 40kDa subunit (ENSG00000124787), score: 0.64 SCLT1sodium channel and clathrin linker 1 (ENSG00000151466), score: 0.87 SEPT12septin 12 (ENSG00000140623), score: 0.6 SERINC4serine incorporator 4 (ENSG00000184716), score: 0.99 SLC13A1solute carrier family 13 (sodium/sulfate symporters), member 1 (ENSG00000081800), score: 0.69 SLC22A4solute carrier family 22 (organic cation/ergothioneine transporter), member 4 (ENSG00000197208), score: 0.61 SLC45A3solute carrier family 45, member 3 (ENSG00000158715), score: 0.67 SLC6A2solute carrier family 6 (neurotransmitter transporter, noradrenalin), member 2 (ENSG00000103546), score: 0.6 SNX1sorting nexin 1 (ENSG00000028528), score: 0.66 SPG21spastic paraplegia 21 (autosomal recessive, Mast syndrome) (ENSG00000090487), score: 0.6 TMEM104transmembrane protein 104 (ENSG00000109066), score: 0.6 TNFSF15tumor necrosis factor (ligand) superfamily, member 15 (ENSG00000181634), score: 0.86 TNNtenascin N (ENSG00000120332), score: 0.7 TRAF2TNF receptor-associated factor 2 (ENSG00000127191), score: 0.6 TRPM1transient receptor potential cation channel, subfamily M, member 1 (ENSG00000134160), score: 0.67 TSKUtsukushi small leucine rich proteoglycan homolog (Xenopus laevis) (ENSG00000182704), score: 0.65 WARS2tryptophanyl tRNA synthetase 2, mitochondrial (ENSG00000116874), score: 0.6 XPCxeroderma pigmentosum, complementation group C (ENSG00000154767), score: 0.73 ZNF618zinc finger protein 618 (ENSG00000157657), score: 0.59 ZNHIT3zinc finger, HIT type 3 (ENSG00000108278), score: -0.62
| Id | species | tissue | sex | individual |
|---|---|---|---|---|
| mml_kd_m_ca1 | mml | kd | m | _ |
| mml_kd_f_ca1 | mml | kd | f | _ |
| mml_lv_f_ca1 | mml | lv | f | _ |
| mml_lv_m_ca1 | mml | lv | m | _ |