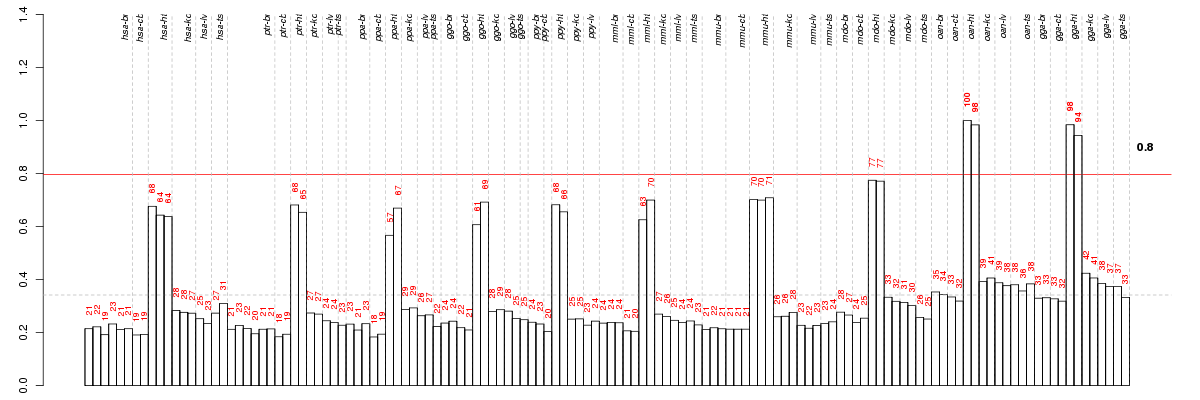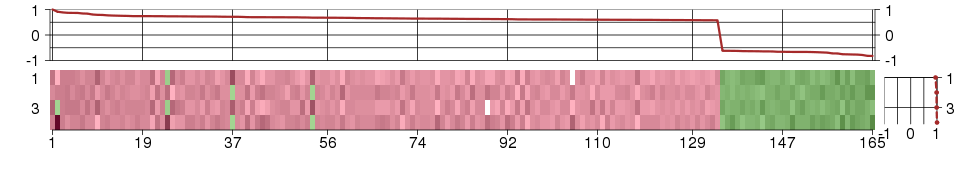

Under-expression is coded with green,
over-expression with red color.

metabolic process
The chemical reactions and pathways, including anabolism and catabolism, by which living organisms transform chemical substances. Metabolic processes typically transform small molecules, but also include macromolecular processes such as DNA repair and replication, and protein synthesis and degradation.
regulation of the force of heart contraction
Any process that modulates the extent of heart contraction, changing the force with which blood is propelled.
heart morphogenesis
The developmental process by which the heart is generated and organized. The heart is a hollow, muscular organ, which, by contracting rhythmically, keeps up the circulation of the blood.
system process
A multicellular organismal process carried out by any of the organs or tissues in an organ system. An organ system is a regularly interacting or interdependent group of organs or tissues that work together to carry out a biological objective.
muscle system process
A organ system process carried out at the level of a muscle. Muscle tissue is composed of contractile cells or fibers.
circulatory system process
A organ system process carried out by any of the organs or tissues of the circulatory system. The circulatory system is an organ system that moves extracellular fluids to and from tissue within a multicellular organism.
heart process
A circulatory system process carried out by the heart. The heart is a hollow, muscular organ, which, by contracting rhythmically, keeps up the circulation of the blood. The heart is a hollow, muscular organ, which, by contracting rhythmically, keeps up the circulation of the blood.
cardiac chamber development
The progression of a cardiac chamber over time, from its formation to the mature structure. A cardiac chamber is an enclosed cavity within the heart.
cardiac chamber morphogenesis
The process by which a cardiac chamber is generated and organized. A cardiac chamber is an enclosed cavity within the heart.
cardiac ventricle morphogenesis
The process by which the cardiac ventricle is generated and organized. A cardiac ventricle receives blood from a cardiac atrium and pumps it out of the heart.
ventricular cardiac muscle tissue development
The process whose specific outcome is the progression of ventricular cardiac muscle over time, from its formation to the mature structure.
cardiac ventricle development
The process whose specific outcome is the progression of a cardiac ventricle over time, from its formation to the mature structure. A cardiac ventricle receives blood from a cardiac atrium and pumps it out of the heart.
muscle contraction
A process whereby force is generated within muscle tissue, resulting in a change in muscle geometry. Force generation involves a chemo-mechanical energy conversion step that is carried out by the actin/myosin complex activity, which generates force through ATP hydrolysis.
nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
Any cellular metabolic process involving nucleobases, nucleosides, nucleotides and nucleic acids.
regulation of nucleotide metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways involving nucleotides.
purine nucleotide metabolic process
The chemical reactions and pathways involving a purine nucleotide, a compound consisting of nucleoside (a purine base linked to a deoxyribose or ribose sugar) esterified with a phosphate moiety at either the 3' or 5'-hydroxyl group of its glycose moiety.
purine nucleotide catabolic process
The chemical reactions and pathways resulting in the breakdown of a purine nucleotide, a compound consisting of nucleoside (a purine base linked to a deoxyribose or ribose sugar) esterified with a phosphate moiety at either the 3' or 5'-hydroxyl group of its glycose moiety.
nucleoside phosphate metabolic process
The chemical reactions and pathways involving any phosphorylated nucleoside.
nitrogen compound metabolic process
The chemical reactions and pathways involving various organic and inorganic nitrogenous compounds; includes nitrogen fixation, nitrification, denitrification, assimilatory/dissimilatory nitrate reduction and the interconversion of nitrogenous organic matter and ammonium.
multicellular organismal development
The biological process whose specific outcome is the progression of a multicellular organism over time from an initial condition (e.g. a zygote or a young adult) to a later condition (e.g. a multicellular animal or an aged adult).
anatomical structure morphogenesis
The process by which anatomical structures are generated and organized. Morphogenesis pertains to the creation of form.
organ morphogenesis
Morphogenesis of an organ. An organ is defined as a tissue or set of tissues that work together to perform a specific function or functions. Morphogenesis is the process by which anatomical structures are generated and organized. Organs are commonly observed as visibly distinct structures, but may also exist as loosely associated clusters of cells that work together to perform a specific function or functions.
tissue development
The process whose specific outcome is the progression of a tissue over time, from its formation to the mature structure.
heart development
The process whose specific outcome is the progression of the heart over time, from its formation to the mature structure. The heart is a hollow, muscular organ, which, by contracting rhythmically, keeps up the circulation of the blood.
muscle organ development
The process whose specific outcome is the progression of the muscle over time, from its formation to the mature structure. The muscle is an organ consisting of a tissue made up of various elongated cells that are specialized to contract and thus to produce movement and mechanical work.
blood circulation
The flow of blood through the body of an animal, enabling the transport of nutrients to the tissues and the removal of waste products.
regulation of heart contraction
Any process that modulates the frequency, rate or extent of heart contraction. Heart contraction is the process by which the heart decreases in volume in a characteristic way to propel blood through the body.
biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
catabolic process
The chemical reactions and pathways resulting in the breakdown of substances, including the breakdown of carbon compounds with the liberation of energy for use by the cell or organism.
nucleotide metabolic process
The chemical reactions and pathways involving a nucleotide, a nucleoside that is esterified with (ortho)phosphate or an oligophosphate at any hydroxyl group on the glycose moiety; may be mono-, di- or triphosphate; this definition includes cyclic nucleotides (nucleoside cyclic phosphates).
nucleotide catabolic process
The chemical reactions and pathways resulting in the breakdown of nucleotides, any nucleoside that is esterified with (ortho)phosphate or an oligophosphate at any hydroxyl group on the glycose moiety; may be mono-, di- or triphosphate; this definition includes cyclic-nucleotides (nucleoside cyclic phosphates).
regulation of catabolic process
Any process that modulates the frequency, rate, or extent of the chemical reactions and pathways resulting in the breakdown of substances.
cellular process
Any process that is carried out at the cellular level, but not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
striated muscle tissue development
The process whose specific outcome is the progression of a striated muscle over time, from its formation to the mature structure. Striated muscle contain fibers that are divided by transverse bands into striations, and cardiac and skeletal muscle are types of striated muscle. Skeletal muscle myoblasts fuse to form myotubes and eventually multinucleated muscle fibers. The fusion of cardiac cells is very rare and can only form binucleate cells.
regulation of nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
Any cellular process that modulates the frequency, rate or extent of the chemical reactions and pathways involving nucleobases, nucleosides, nucleotides and nucleic acids.
regulation of metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways within a cell or an organism.
regulation of nucleotide catabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways resulting in the breakdown of nucleotides.
regulation of cellular metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways by which individual cells transform chemical substances.
regulation of cellular catabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways resulting in the breakdown of substances, carried out by individual cells.
multicellular organismal process
Any biological process, occurring at the level of a multicellular organism, pertinent to its function.
developmental process
A biological process whose specific outcome is the progression of an integrated living unit: an anatomical structure (which may be a subcellular structure, cell, tissue, or organ), or organism over time from an initial condition to a later condition.
regulation of purine nucleotide catabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways resulting in the breakdown of purine nucleotides.
cellular nitrogen compound metabolic process
The chemical reactions and pathways involving various organic and inorganic nitrogenous compounds, as carried out by individual cells.
nucleobase, nucleoside, nucleotide and nucleic acid catabolic process
The chemical reactions and pathways resulting in the breakdown of nucleobases, nucleosides, nucleotides and nucleic acids.
nucleobase, nucleoside and nucleotide catabolic process
The chemical reactions and pathways resulting in the breakdown of nucleobases, nucleosides and nucleotides.
regulation of ATPase activity
Any process that modulates the rate of ATP hydrolysis by an ATPase.
regulation of system process
Any process that modulates the frequency, rate or extent of a system process, a multicellular organismal process carried out by any of the organs or tissues in an organ system.
cellular metabolic process
The chemical reactions and pathways by which individual cells transform chemical substances.
primary metabolic process
The chemical reactions and pathways involving those compounds which are formed as a part of the normal anabolic and catabolic processes. These processes take place in most, if not all, cells of the organism.
cellular catabolic process
The chemical reactions and pathways resulting in the breakdown of substances, carried out by individual cells.
cellular nitrogen compound catabolic process
The chemical reactions and pathways resulting in the breakdown of organic and inorganic nitrogenous compounds.
small molecule metabolic process
The chemical reactions and pathways involving small molecules, any monomeric molecule of small relative molecular mass.
small molecule catabolic process
The chemical reactions and pathways resulting in the breakdown of small molecules, any monomeric molecule of small relative molecular mass.
heterocycle metabolic process
The chemical reactions and pathways involving heterocyclic compounds, those with a cyclic molecular structure and at least two different atoms in the ring (or rings).
heterocycle catabolic process
The chemical reactions and pathways resulting in the breakdown of heterocyclic compounds, those with a cyclic molecular structure and at least two different atoms in the ring (or rings).
organ development
Development of a tissue or tissues that work together to perform a specific function or functions. Development pertains to the process whose specific outcome is the progression of a structure over time, from its formation to the mature structure. Organs are commonly observed as visibly distinct structures, but may also exist as loosely associated clusters of cells that work together to perform a specific function or functions.
tissue morphogenesis
The process by which the anatomical structures of a tissue are generated and organized. Morphogenesis pertains to the creation of form.
system development
The process whose specific outcome is the progression of an organismal system over time, from its formation to the mature structure. A system is a regularly interacting or interdependent group of organs or tissues that work together to carry out a given biological process.
cardiac muscle tissue development
The process whose specific outcome is the progression of cardiac muscle over time, from its formation to the mature structure.
anatomical structure development
The biological process whose specific outcome is the progression of an anatomical structure from an initial condition to its mature state. This process begins with the formation of the structure and ends with the mature structure, whatever form that may be including its natural destruction. An anatomical structure is any biological entity that occupies space and is distinguished from its surroundings. Anatomical structures can be macroscopic such as a carpel, or microscopic such as an acrosome.
regulation of biological process
Any process that modulates the frequency, rate or extent of a biological process. Biological processes are regulated by many means; examples include the control of gene expression, protein modification or interaction with a protein or substrate molecule.
regulation of catalytic activity
Any process that modulates the activity of an enzyme.
regulation of cellular process
Any process that modulates the frequency, rate or extent of a cellular process, any of those that are carried out at the cellular level, but are not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
regulation of nitrogen compound metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways involving nitrogen or nitrogenous compounds.
regulation of multicellular organismal process
Any process that modulates the frequency, rate or extent of a multicellular organismal process, the processes pertinent to the function of a multicellular organism above the cellular level; includes the integrated processes of tissues and organs.
regulation of hydrolase activity
Any process that modulates the frequency, rate or extent of hydrolase activity, the catalysis of the hydrolysis of various bonds, e.g. C-O, C-N, C-C, phosphoric anhydride bonds, etc. Hydrolase is the systematic name for any enzyme of EC class 3.
cardiac muscle tissue morphogenesis
The process by which the anatomical structures of cardiac muscle tissue are generated and organized. Morphogenesis pertains to the creation of form.
ventricular cardiac muscle tissue morphogenesis
The process by which the anatomical structures of cardiac ventricle muscle is generated and organized. Morphogenesis pertains to the creation of form.
nucleobase, nucleoside and nucleotide metabolic process
The cellular chemical reactions and pathways involving nucleobases, nucleosides and nucleotides.
heart contraction
The multicellular organismal process by which the heart decreases in volume in a characteristic way to propel blood through the body.
muscle tissue morphogenesis
The process by which the anatomical structures of muscle tissue are generated and organized. Muscle tissue consists of a set of cells that are part of an organ and carry out a contractive function. Morphogenesis pertains to the creation of form.
muscle tissue development
The progression of muscle tissue over time, from its initial formation to its mature state. Muscle tissue is a contractile tissue made up of actin and myosin fibers.
muscle structure development
The progression of a muscle structure over time, from its formation to its mature state. Muscle structures are contractile cells, tissues or organs that are found in multicellular organisms.
biological regulation
Any process that modulates the frequency, rate or extent of any biological process, quality or function.
regulation of biological quality
Any process that modulates the frequency, rate or extent of a biological quality. A biological quality is a measurable attribute of an organism or part of an organism, such as size, mass, shape, color, etc.
regulation of molecular function
Any process that modulates the frequency, rate or extent of a molecular function, an elemental biological activity occurring at the molecular level, such as catalysis or binding.
regulation of primary metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways within a cell or an organism involving those compounds formed as a part of the normal anabolic and catabolic processes. These processes take place in most, if not all, cells of the organism.
all
NA
cellular metabolic process
The chemical reactions and pathways by which individual cells transform chemical substances.
multicellular organismal development
The biological process whose specific outcome is the progression of a multicellular organism over time from an initial condition (e.g. a zygote or a young adult) to a later condition (e.g. a multicellular animal or an aged adult).
regulation of metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways within a cell or an organism.
regulation of cellular process
Any process that modulates the frequency, rate or extent of a cellular process, any of those that are carried out at the cellular level, but are not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
regulation of multicellular organismal process
Any process that modulates the frequency, rate or extent of a multicellular organismal process, the processes pertinent to the function of a multicellular organism above the cellular level; includes the integrated processes of tissues and organs.
regulation of biological process
Any process that modulates the frequency, rate or extent of a biological process. Biological processes are regulated by many means; examples include the control of gene expression, protein modification or interaction with a protein or substrate molecule.
regulation of catabolic process
Any process that modulates the frequency, rate, or extent of the chemical reactions and pathways resulting in the breakdown of substances.
regulation of nitrogen compound metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways involving nitrogen or nitrogenous compounds.
regulation of cellular metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways by which individual cells transform chemical substances.
cellular nitrogen compound metabolic process
The chemical reactions and pathways involving various organic and inorganic nitrogenous compounds, as carried out by individual cells.
cellular catabolic process
The chemical reactions and pathways resulting in the breakdown of substances, carried out by individual cells.
regulation of primary metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways within a cell or an organism involving those compounds formed as a part of the normal anabolic and catabolic processes. These processes take place in most, if not all, cells of the organism.
small molecule catabolic process
The chemical reactions and pathways resulting in the breakdown of small molecules, any monomeric molecule of small relative molecular mass.
regulation of cellular metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways by which individual cells transform chemical substances.
regulation of system process
Any process that modulates the frequency, rate or extent of a system process, a multicellular organismal process carried out by any of the organs or tissues in an organ system.
anatomical structure morphogenesis
The process by which anatomical structures are generated and organized. Morphogenesis pertains to the creation of form.
system development
The process whose specific outcome is the progression of an organismal system over time, from its formation to the mature structure. A system is a regularly interacting or interdependent group of organs or tissues that work together to carry out a given biological process.
regulation of catalytic activity
Any process that modulates the activity of an enzyme.
nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
Any cellular metabolic process involving nucleobases, nucleosides, nucleotides and nucleic acids.
regulation of cellular catabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways resulting in the breakdown of substances, carried out by individual cells.
cellular nitrogen compound catabolic process
The chemical reactions and pathways resulting in the breakdown of organic and inorganic nitrogenous compounds.
regulation of nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
Any cellular process that modulates the frequency, rate or extent of the chemical reactions and pathways involving nucleobases, nucleosides, nucleotides and nucleic acids.
regulation of cellular catabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways resulting in the breakdown of substances, carried out by individual cells.
regulation of nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
Any cellular process that modulates the frequency, rate or extent of the chemical reactions and pathways involving nucleobases, nucleosides, nucleotides and nucleic acids.
heterocycle catabolic process
The chemical reactions and pathways resulting in the breakdown of heterocyclic compounds, those with a cyclic molecular structure and at least two different atoms in the ring (or rings).
regulation of nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
Any cellular process that modulates the frequency, rate or extent of the chemical reactions and pathways involving nucleobases, nucleosides, nucleotides and nucleic acids.
nucleobase, nucleoside and nucleotide metabolic process
The cellular chemical reactions and pathways involving nucleobases, nucleosides and nucleotides.
nucleobase, nucleoside and nucleotide catabolic process
The chemical reactions and pathways resulting in the breakdown of nucleobases, nucleosides and nucleotides.
organ development
Development of a tissue or tissues that work together to perform a specific function or functions. Development pertains to the process whose specific outcome is the progression of a structure over time, from its formation to the mature structure. Organs are commonly observed as visibly distinct structures, but may also exist as loosely associated clusters of cells that work together to perform a specific function or functions.
tissue morphogenesis
The process by which the anatomical structures of a tissue are generated and organized. Morphogenesis pertains to the creation of form.
cardiac chamber morphogenesis
The process by which a cardiac chamber is generated and organized. A cardiac chamber is an enclosed cavity within the heart.
tissue morphogenesis
The process by which the anatomical structures of a tissue are generated and organized. Morphogenesis pertains to the creation of form.
organ morphogenesis
Morphogenesis of an organ. An organ is defined as a tissue or set of tissues that work together to perform a specific function or functions. Morphogenesis is the process by which anatomical structures are generated and organized. Organs are commonly observed as visibly distinct structures, but may also exist as loosely associated clusters of cells that work together to perform a specific function or functions.
tissue development
The process whose specific outcome is the progression of a tissue over time, from its formation to the mature structure.
muscle organ development
The process whose specific outcome is the progression of the muscle over time, from its formation to the mature structure. The muscle is an organ consisting of a tissue made up of various elongated cells that are specialized to contract and thus to produce movement and mechanical work.
nucleobase, nucleoside, nucleotide and nucleic acid catabolic process
The chemical reactions and pathways resulting in the breakdown of nucleobases, nucleosides, nucleotides and nucleic acids.
purine nucleotide catabolic process
The chemical reactions and pathways resulting in the breakdown of a purine nucleotide, a compound consisting of nucleoside (a purine base linked to a deoxyribose or ribose sugar) esterified with a phosphate moiety at either the 3' or 5'-hydroxyl group of its glycose moiety.
nucleobase, nucleoside and nucleotide catabolic process
The chemical reactions and pathways resulting in the breakdown of nucleobases, nucleosides and nucleotides.
heart contraction
The multicellular organismal process by which the heart decreases in volume in a characteristic way to propel blood through the body.
regulation of the force of heart contraction
Any process that modulates the extent of heart contraction, changing the force with which blood is propelled.
cardiac chamber morphogenesis
The process by which a cardiac chamber is generated and organized. A cardiac chamber is an enclosed cavity within the heart.
cardiac muscle tissue morphogenesis
The process by which the anatomical structures of cardiac muscle tissue are generated and organized. Morphogenesis pertains to the creation of form.
cardiac ventricle morphogenesis
The process by which the cardiac ventricle is generated and organized. A cardiac ventricle receives blood from a cardiac atrium and pumps it out of the heart.
heart morphogenesis
The developmental process by which the heart is generated and organized. The heart is a hollow, muscular organ, which, by contracting rhythmically, keeps up the circulation of the blood.
cardiac chamber development
The progression of a cardiac chamber over time, from its formation to the mature structure. A cardiac chamber is an enclosed cavity within the heart.
muscle tissue development
The progression of muscle tissue over time, from its initial formation to its mature state. Muscle tissue is a contractile tissue made up of actin and myosin fibers.
regulation of nucleotide catabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways resulting in the breakdown of nucleotides.
regulation of purine nucleotide catabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways resulting in the breakdown of purine nucleotides.
purine nucleotide catabolic process
The chemical reactions and pathways resulting in the breakdown of a purine nucleotide, a compound consisting of nucleoside (a purine base linked to a deoxyribose or ribose sugar) esterified with a phosphate moiety at either the 3' or 5'-hydroxyl group of its glycose moiety.
regulation of nucleotide catabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways resulting in the breakdown of nucleotides.
regulation of nucleotide metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways involving nucleotides.
purine nucleotide metabolic process
The chemical reactions and pathways involving a purine nucleotide, a compound consisting of nucleoside (a purine base linked to a deoxyribose or ribose sugar) esterified with a phosphate moiety at either the 3' or 5'-hydroxyl group of its glycose moiety.
nucleotide catabolic process
The chemical reactions and pathways resulting in the breakdown of nucleotides, any nucleoside that is esterified with (ortho)phosphate or an oligophosphate at any hydroxyl group on the glycose moiety; may be mono-, di- or triphosphate; this definition includes cyclic-nucleotides (nucleoside cyclic phosphates).
regulation of heart contraction
Any process that modulates the frequency, rate or extent of heart contraction. Heart contraction is the process by which the heart decreases in volume in a characteristic way to propel blood through the body.
ventricular cardiac muscle tissue morphogenesis
The process by which the anatomical structures of cardiac ventricle muscle is generated and organized. Morphogenesis pertains to the creation of form.
cardiac muscle tissue development
The process whose specific outcome is the progression of cardiac muscle over time, from its formation to the mature structure.
cardiac muscle tissue morphogenesis
The process by which the anatomical structures of cardiac muscle tissue are generated and organized. Morphogenesis pertains to the creation of form.
regulation of ATPase activity
Any process that modulates the rate of ATP hydrolysis by an ATPase.
ventricular cardiac muscle tissue morphogenesis
The process by which the anatomical structures of cardiac ventricle muscle is generated and organized. Morphogenesis pertains to the creation of form.

intracellular
The living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
contractile fiber
Fibers, composed of actin, myosin, and associated proteins, found in cells of smooth or striated muscle.
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
cell
The basic structural and functional unit of all organisms. Includes the plasma membrane and any external encapsulating structures such as the cell wall and cell envelope.
cytoplasm
All of the contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures.
myofibril
The contractile element of skeletal and cardiac muscle; a long, highly organized bundle of actin, myosin, and other proteins that contracts by a sliding filament mechanism.
sarcomere
The repeating unit of a myofibril in a muscle cell, composed of an array of overlapping thick and thin filaments between two adjacent Z discs.
Z disc
Platelike region of a muscle sarcomere to which the plus ends of actin filaments are attached.
A band
The dark-staining region of a sarcomere, in which myosin thick filaments are present; the center is traversed by the paler H zone, which in turn contains the M line.
I band
A region of a sarcomere that appears as a light band on each side of the Z disc, comprising a region of the sarcomere where thin (actin) filaments are not overlapped by thick (myosin) filaments; contains actin, troponin, and tropomyosin; each sarcomere includes half of an I band at each end.
organelle
Organized structure of distinctive morphology and function. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton, and prokaryotic structures such as anammoxosomes and pirellulosomes. Excludes the plasma membrane.
non-membrane-bounded organelle
Organized structure of distinctive morphology and function, not bounded by a lipid bilayer membrane. Includes ribosomes, the cytoskeleton and chromosomes.
intracellular organelle
Organized structure of distinctive morphology and function, occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton. Excludes the plasma membrane.
intracellular non-membrane-bounded organelle
Organized structure of distinctive morphology and function, not bounded by a lipid bilayer membrane and occurring within the cell. Includes ribosomes, the cytoskeleton and chromosomes.
organelle part
Any constituent part of an organelle, an organized structure of distinctive morphology and function. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton, but excludes the plasma membrane.
intracellular part
Any constituent part of the living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
cytoplasmic part
Any constituent part of the cytoplasm, all of the contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures.
contractile fiber part
Any constituent part of a contractile fiber, a fiber composed of actin, myosin, and associated proteins, found in cells of smooth or striated muscle.
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
all
NA
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
organelle part
Any constituent part of an organelle, an organized structure of distinctive morphology and function. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton, but excludes the plasma membrane.
intracellular non-membrane-bounded organelle
Organized structure of distinctive morphology and function, not bounded by a lipid bilayer membrane and occurring within the cell. Includes ribosomes, the cytoskeleton and chromosomes.
intracellular part
Any constituent part of the living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
intracellular organelle
Organized structure of distinctive morphology and function, occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton. Excludes the plasma membrane.
A band
The dark-staining region of a sarcomere, in which myosin thick filaments are present; the center is traversed by the paler H zone, which in turn contains the M line.
I band
A region of a sarcomere that appears as a light band on each side of the Z disc, comprising a region of the sarcomere where thin (actin) filaments are not overlapped by thick (myosin) filaments; contains actin, troponin, and tropomyosin; each sarcomere includes half of an I band at each end.
Z disc
Platelike region of a muscle sarcomere to which the plus ends of actin filaments are attached.
cytoplasmic part
Any constituent part of the cytoplasm, all of the contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures.
contractile fiber
Fibers, composed of actin, myosin, and associated proteins, found in cells of smooth or striated muscle.
contractile fiber part
Any constituent part of a contractile fiber, a fiber composed of actin, myosin, and associated proteins, found in cells of smooth or striated muscle.
contractile fiber part
Any constituent part of a contractile fiber, a fiber composed of actin, myosin, and associated proteins, found in cells of smooth or striated muscle.
sarcomere
The repeating unit of a myofibril in a muscle cell, composed of an array of overlapping thick and thin filaments between two adjacent Z discs.

protein binding
Interacting selectively and non-covalently with any protein or protein complex (a complex of two or more proteins that may include other nonprotein molecules).
molecular_function
Elemental activities, such as catalysis or binding, describing the actions of a gene product at the molecular level. A given gene product may exhibit one or more molecular functions.
binding
The selective, non-covalent, often stoichiometric, interaction of a molecule with one or more specific sites on another molecule.
cytoskeletal protein binding
Interacting selectively and non-covalently with any protein component of any cytoskeleton (actin, microtubule, or intermediate filament cytoskeleton).
titin binding
Interacting selectively and non-covalently with titin, any of a family of giant proteins found in striated and smooth muscle. In striated muscle, single titin molecules span half the sarcomere, with their N- and C-termini in the Z-disc and M-line, respectively.
all
NA
| Id | Pvalue | ExpCount | Count | Size | Term |
|---|---|---|---|---|---|
| 05410 | 6.749e-03 | 1.169 | 7 | 34 | Hypertrophic cardiomyopathy (HCM) |
| 04260 | 8.431e-03 | 0.8595 | 6 | 25 | Cardiac muscle contraction |
| 05414 | 3.201e-02 | 1.169 | 6 | 34 | Dilated cardiomyopathy |
ABHD12abhydrolase domain containing 12 (ENSG00000100997), score: -0.66 ACAD8acyl-CoA dehydrogenase family, member 8 (ENSG00000151498), score: 0.6 ACAD9acyl-CoA dehydrogenase family, member 9 (ENSG00000177646), score: 0.65 ACTN2actinin, alpha 2 (ENSG00000077522), score: 0.6 ADAadenosine deaminase (ENSG00000196839), score: 0.6 ADAMTSL5ADAMTS-like 5 (ENSG00000185761), score: 0.58 ADAT1adenosine deaminase, tRNA-specific 1 (ENSG00000065457), score: 0.6 AHCYL1adenosylhomocysteinase-like 1 (ENSG00000168710), score: -0.75 AIG1androgen-induced 1 (ENSG00000146416), score: -0.72 AIMP1aminoacyl tRNA synthetase complex-interacting multifunctional protein 1 (ENSG00000164022), score: 0.62 AMPD1adenosine monophosphate deaminase 1 (ENSG00000116748), score: 0.91 APOBEC2apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 2 (ENSG00000124701), score: 0.74 ARFGEF2ADP-ribosylation factor guanine nucleotide-exchange factor 2 (brefeldin A-inhibited) (ENSG00000124198), score: -0.82 ARL14ADP-ribosylation factor-like 14 (ENSG00000179674), score: 0.63 ARL5BADP-ribosylation factor-like 5B (ENSG00000165997), score: 0.59 ATP1B4ATPase, Na+/K+ transporting, beta 4 polypeptide (ENSG00000101892), score: 0.74 ATP7AATPase, Cu++ transporting, alpha polypeptide (ENSG00000165240), score: 0.6 B4GALT5UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 5 (ENSG00000158470), score: -0.68 BCKDHBbranched chain keto acid dehydrogenase E1, beta polypeptide (ENSG00000083123), score: 0.59 BCL2L13BCL2-like 13 (apoptosis facilitator) (ENSG00000099968), score: 0.84 BEST3bestrophin 3 (ENSG00000127325), score: 0.79 BSDC1BSD domain containing 1 (ENSG00000160058), score: -0.77 C12orf45chromosome 12 open reading frame 45 (ENSG00000151131), score: 0.68 C16orf7chromosome 16 open reading frame 7 (ENSG00000075399), score: -0.64 C1QTNF7C1q and tumor necrosis factor related protein 7 (ENSG00000163145), score: 0.87 C6orf142chromosome 6 open reading frame 142 (ENSG00000146147), score: 0.63 CCNIcyclin I (ENSG00000118816), score: 0.69 CDCP2CUB domain containing protein 2 (ENSG00000157211), score: 0.68 CDK6cyclin-dependent kinase 6 (ENSG00000105810), score: 0.6 CHRNA5cholinergic receptor, nicotinic, alpha 5 (ENSG00000169684), score: 0.69 CLEC3AC-type lectin domain family 3, member A (ENSG00000166509), score: 0.71 CLMNcalmin (calponin-like, transmembrane) (ENSG00000165959), score: -0.62 CLPXClpX caseinolytic peptidase X homolog (E. coli) (ENSG00000166855), score: 0.59 CLTAclathrin, light chain A (ENSG00000122705), score: -0.82 CMYA5cardiomyopathy associated 5 (ENSG00000164309), score: 0.75 COMMD2COMM domain containing 2 (ENSG00000114744), score: 0.59 COPS7ACOP9 constitutive photomorphogenic homolog subunit 7A (Arabidopsis) (ENSG00000111652), score: 0.63 COQ9coenzyme Q9 homolog (S. cerevisiae) (ENSG00000088682), score: 0.63 CSRP3cysteine and glycine-rich protein 3 (cardiac LIM protein) (ENSG00000129170), score: 0.68 CYSLTR1cysteinyl leukotriene receptor 1 (ENSG00000173198), score: 0.64 DCAF6DDB1 and CUL4 associated factor 6 (ENSG00000143164), score: 0.72 DCTN6dynactin 6 (ENSG00000104671), score: 0.59 DDAH1dimethylarginine dimethylaminohydrolase 1 (ENSG00000153904), score: -0.62 DHCR77-dehydrocholesterol reductase (ENSG00000172893), score: -0.69 DLDdihydrolipoamide dehydrogenase (ENSG00000091140), score: 0.63 DNTTIP1deoxynucleotidyltransferase, terminal, interacting protein 1 (ENSG00000101457), score: 0.7 DSCR3Down syndrome critical region gene 3 (ENSG00000157538), score: 0.64 DUSP27dual specificity phosphatase 27 (putative) (ENSG00000198842), score: 0.58 EDNRAendothelin receptor type A (ENSG00000151617), score: 0.7 EHD4EH-domain containing 4 (ENSG00000103966), score: 0.61 EIF3Deukaryotic translation initiation factor 3, subunit D (ENSG00000100353), score: 0.72 ELP2elongation protein 2 homolog (S. cerevisiae) (ENSG00000134759), score: -0.62 F2RL2coagulation factor II (thrombin) receptor-like 2 (ENSG00000164220), score: 0.73 F8coagulation factor VIII, procoagulant component (ENSG00000185010), score: 0.7 FBP2fructose-1,6-bisphosphatase 2 (ENSG00000130957), score: 0.71 FBXO40F-box protein 40 (ENSG00000163833), score: 0.65 FGF6fibroblast growth factor 6 (ENSG00000111241), score: 0.73 FGFR3fibroblast growth factor receptor 3 (ENSG00000068078), score: -0.66 FHL2four and a half LIM domains 2 (ENSG00000115641), score: 0.67 FHL3four and a half LIM domains 3 (ENSG00000183386), score: 0.8 FIGFc-fos induced growth factor (vascular endothelial growth factor D) (ENSG00000165197), score: 0.74 FILIP1filamin A interacting protein 1 (ENSG00000118407), score: 0.73 FSD2fibronectin type III and SPRY domain containing 2 (ENSG00000186628), score: 0.66 FZD2frizzled homolog 2 (Drosophila) (ENSG00000180340), score: 0.73 GATA5GATA binding protein 5 (ENSG00000130700), score: 0.84 GATMglycine amidinotransferase (L-arginine:glycine amidinotransferase) (ENSG00000171766), score: -0.63 GPR1G protein-coupled receptor 1 (ENSG00000183671), score: 0.7 HIBCH3-hydroxyisobutyryl-CoA hydrolase (ENSG00000198130), score: 0.65 HIF1ANhypoxia inducible factor 1, alpha subunit inhibitor (ENSG00000166135), score: 0.62 HMGXB4HMG box domain containing 4 (ENSG00000100281), score: 0.69 HSPA13heat shock protein 70kDa family, member 13 (ENSG00000155304), score: -0.66 IL15interleukin 15 (ENSG00000164136), score: 0.61 JOSD1Josephin domain containing 1 (ENSG00000100221), score: -0.67 KBTBD10kelch repeat and BTB (POZ) domain containing 10 (ENSG00000239474), score: 0.64 KCNJ2potassium inwardly-rectifying channel, subfamily J, member 2 (ENSG00000123700), score: 0.7 KDELC1KDEL (Lys-Asp-Glu-Leu) containing 1 (ENSG00000134901), score: 0.59 KIAA0907KIAA0907 (ENSG00000132680), score: 0.67 KIAA1598KIAA1598 (ENSG00000187164), score: -0.72 KLHDC2kelch domain containing 2 (ENSG00000165516), score: 0.63 KLHL31kelch-like 31 (Drosophila) (ENSG00000124743), score: 0.75 LAMA4laminin, alpha 4 (ENSG00000112769), score: 0.58 LMOD2leiomodin 2 (cardiac) (ENSG00000170807), score: 0.64 LOC100291671similar to SH3-binding domain and glutamic acid-rich protein (ENSG00000185437), score: 0.61 LRP11low density lipoprotein receptor-related protein 11 (ENSG00000120256), score: -0.67 LRRC10leucine rich repeat containing 10 (ENSG00000198812), score: 0.77 LRRC17leucine rich repeat containing 17 (ENSG00000128606), score: 0.6 LRRC2leucine rich repeat containing 2 (ENSG00000163827), score: 0.68 LRRC38leucine rich repeat containing 38 (ENSG00000162494), score: 0.61 MAD2L1BPMAD2L1 binding protein (ENSG00000124688), score: -0.66 MAK16MAK16 homolog (S. cerevisiae) (ENSG00000198042), score: 0.7 MAP3K4mitogen-activated protein kinase kinase kinase 4 (ENSG00000085511), score: 0.69 MAP7microtubule-associated protein 7 (ENSG00000135525), score: -0.78 MDH2malate dehydrogenase 2, NAD (mitochondrial) (ENSG00000146701), score: 0.59 MEOX2mesenchyme homeobox 2 (ENSG00000106511), score: 0.71 MRPL50mitochondrial ribosomal protein L50 (ENSG00000136897), score: 0.63 MRPS27mitochondrial ribosomal protein S27 (ENSG00000113048), score: 0.72 MYBPC3myosin binding protein C, cardiac (ENSG00000134571), score: 0.68 MYL3myosin, light chain 3, alkali; ventricular, skeletal, slow (ENSG00000160808), score: 0.66 MYLIPmyosin regulatory light chain interacting protein (ENSG00000007944), score: 0.58 MYLK3myosin light chain kinase 3 (ENSG00000140795), score: 0.69 MYOCDmyocardin (ENSG00000141052), score: 0.87 MYOTmyotilin (ENSG00000120729), score: 1 MYOZ2myozenin 2 (ENSG00000172399), score: 0.64 NDUFA12NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 12 (ENSG00000184752), score: 0.65 NDUFS3NADH dehydrogenase (ubiquinone) Fe-S protein 3, 30kDa (NADH-coenzyme Q reductase) (ENSG00000213619), score: 0.66 NKIRAS2NFKB inhibitor interacting Ras-like 2 (ENSG00000168256), score: 0.61 NSDHLNAD(P) dependent steroid dehydrogenase-like (ENSG00000147383), score: -0.64 OGDHoxoglutarate (alpha-ketoglutarate) dehydrogenase (lipoamide) (ENSG00000105953), score: 0.59 OTUD1OTU domain containing 1 (ENSG00000165312), score: 0.6 OXSR1oxidative-stress responsive 1 (ENSG00000172939), score: 0.61 PARNpoly(A)-specific ribonuclease (deadenylation nuclease) (ENSG00000140694), score: -0.63 PDE5Aphosphodiesterase 5A, cGMP-specific (ENSG00000138735), score: 0.72 PDLIM5PDZ and LIM domain 5 (ENSG00000163110), score: 0.64 PDRG1p53 and DNA-damage regulated 1 (ENSG00000088356), score: 0.61 PLEKHA7pleckstrin homology domain containing, family A member 7 (ENSG00000166689), score: 0.64 PLNphospholamban (ENSG00000198523), score: 0.72 PM20D2peptidase M20 domain containing 2 (ENSG00000146281), score: 0.7 POLR2Fpolymerase (RNA) II (DNA directed) polypeptide F (ENSG00000100142), score: 0.72 PPP1R13Bprotein phosphatase 1, regulatory (inhibitor) subunit 13B (ENSG00000088808), score: 0.61 PRKAB2protein kinase, AMP-activated, beta 2 non-catalytic subunit (ENSG00000131791), score: 0.6 PRKAG2protein kinase, AMP-activated, gamma 2 non-catalytic subunit (ENSG00000106617), score: 0.64 PSMA3proteasome (prosome, macropain) subunit, alpha type, 3 (ENSG00000100567), score: 0.64 PTPN1protein tyrosine phosphatase, non-receptor type 1 (ENSG00000196396), score: -0.65 PTRFpolymerase I and transcript release factor (ENSG00000177469), score: 0.68 QPCTLglutaminyl-peptide cyclotransferase-like (ENSG00000011478), score: 0.62 RAB10RAB10, member RAS oncogene family (ENSG00000084733), score: 0.58 RAB19RAB19, member RAS oncogene family (ENSG00000146955), score: 0.66 RBM20RNA binding motif protein 20 (ENSG00000203867), score: 0.67 RBM24RNA binding motif protein 24 (ENSG00000112183), score: 0.61 RDXradixin (ENSG00000137710), score: 0.6 RGS5regulator of G-protein signaling 5 (ENSG00000143248), score: 0.61 RNF34ring finger protein 34 (ENSG00000170633), score: -0.76 RORARAR-related orphan receptor A (ENSG00000069667), score: -0.64 SAV1salvador homolog 1 (Drosophila) (ENSG00000151748), score: 0.65 SIKE1suppressor of IKBKE 1 (ENSG00000052723), score: 0.58 SLC20A1solute carrier family 20 (phosphate transporter), member 1 (ENSG00000144136), score: -0.66 SLC8A1solute carrier family 8 (sodium/calcium exchanger), member 1 (ENSG00000183023), score: 0.61 SMPXsmall muscle protein, X-linked (ENSG00000091482), score: 0.73 SMYD1SET and MYND domain containing 1 (ENSG00000115593), score: 0.74 SOD2superoxide dismutase 2, mitochondrial (ENSG00000112096), score: 0.66 SSX2IPsynovial sarcoma, X breakpoint 2 interacting protein (ENSG00000117155), score: -0.64 ST8SIA2ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 2 (ENSG00000140557), score: 0.74 ST8SIA4ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 4 (ENSG00000113532), score: 0.58 STK39serine threonine kinase 39 (ENSG00000198648), score: -0.62 TAB3TGF-beta activated kinase 1/MAP3K7 binding protein 3 (ENSG00000157625), score: 0.6 TBX18T-box 18 (ENSG00000112837), score: 0.61 TBX20T-box 20 (ENSG00000164532), score: 0.72 TBX5T-box 5 (ENSG00000089225), score: 0.89 TCF12transcription factor 12 (ENSG00000140262), score: 0.76 TEAD4TEA domain family member 4 (ENSG00000197905), score: 0.73 TECRLtrans-2,3-enoyl-CoA reductase-like (ENSG00000205678), score: 0.74 TIMM10translocase of inner mitochondrial membrane 10 homolog (yeast) (ENSG00000134809), score: 0.74 TMEM60transmembrane protein 60 (ENSG00000135211), score: 0.7 TMEM9transmembrane protein 9 (ENSG00000116857), score: -0.68 TMOD1tropomodulin 1 (ENSG00000136842), score: 0.6 TNNC1troponin C type 1 (slow) (ENSG00000114854), score: 0.59 TNNT2troponin T type 2 (cardiac) (ENSG00000118194), score: 0.79 TRIM55tripartite motif-containing 55 (ENSG00000147573), score: 0.87 TRIM63tripartite motif-containing 63 (ENSG00000158022), score: 0.76 TSG101tumor susceptibility gene 101 (ENSG00000074319), score: 0.62 TXLNBtaxilin beta (ENSG00000164440), score: 0.61 UBA2ubiquitin-like modifier activating enzyme 2 (ENSG00000126261), score: -0.66 UNC119unc-119 homolog (C. elegans) (ENSG00000109103), score: -0.76 UQCRC2ubiquinol-cytochrome c reductase core protein II (ENSG00000140740), score: 0.66 YIPF1Yip1 domain family, member 1 (ENSG00000058799), score: 0.63
| Id | species | tissue | sex | individual |
|---|---|---|---|---|
| gga_ht_f_ca1 | gga | ht | f | _ |
| oan_ht_f_ca1 | oan | ht | f | _ |
| gga_ht_m_ca1 | gga | ht | m | _ |
| oan_ht_m_ca1 | oan | ht | m | _ |