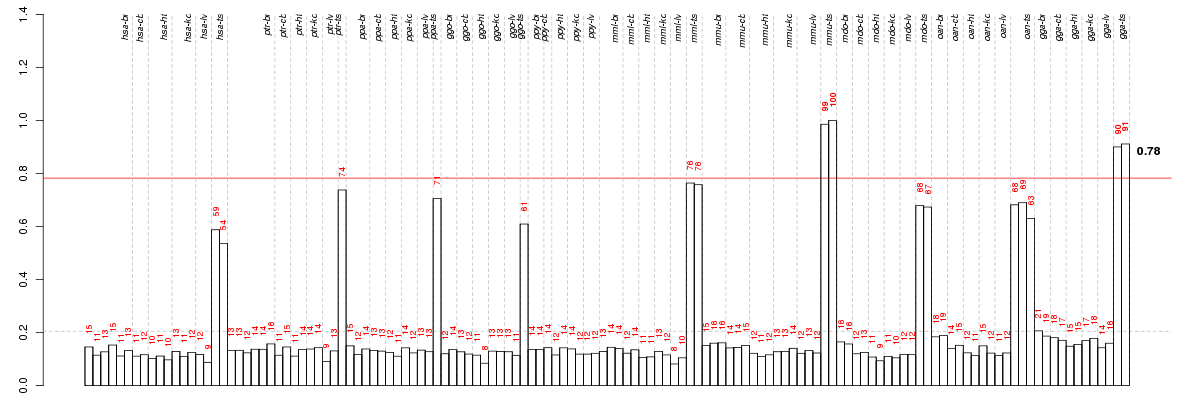

Under-expression is coded with green,
over-expression with red color.

reproduction
The production by an organism of new individuals that contain some portion of their genetic material inherited from that organism.
chromosome segregation
The process by which genetic material, in the form of chromosomes, is organized into specific structures and then physically separated and apportioned to two or more sets.
chromosome organization
A process that is carried out at the cellular level that results in the assembly, arrangement of constituent parts, or disassembly of chromosomes, structures composed of a very long molecule of DNA and associated proteins that carries hereditary information.
M phase of mitotic cell cycle
A cell cycle process comprising the steps by which a cell progresses through M phase, the part of the mitotic cell cycle during which mitosis takes place.
microtubule cytoskeleton organization
A process that is carried out at the cellular level which results in the assembly, arrangement of constituent parts, or disassembly of cytoskeletal structures comprising microtubules and their associated proteins.
mitotic cell cycle
Progression through the phases of the mitotic cell cycle, the most common eukaryotic cell cycle, which canonically comprises four successive phases called G1, S, G2, and M and includes replication of the genome and the subsequent segregation of chromosomes into daughter cells. In some variant cell cycles nuclear replication or nuclear division may not be followed by cell division, or G1 and G2 phases may be absent.
M phase
A cell cycle process comprising the steps by which a cell progresses through M phase, the part of the cell cycle comprising nuclear division.
nuclear division
A process by which a cell nucleus is divided into two nuclei, with DNA and other nuclear contents distributed between the daughter nuclei.
metabolic process
The chemical reactions and pathways, including anabolism and catabolism, by which living organisms transform chemical substances. Metabolic processes typically transform small molecules, but also include macromolecular processes such as DNA repair and replication, and protein synthesis and degradation.
regulation of cell cycle
Any process that modulates the rate or extent of progression through the cell cycle.
cytoskeleton organization
A process that is carried out at the cellular level which results in the assembly, arrangement of constituent parts, or disassembly of cytoskeletal structures.
nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
Any cellular metabolic process involving nucleobases, nucleosides, nucleotides and nucleic acids.
nitrogen compound metabolic process
The chemical reactions and pathways involving various organic and inorganic nitrogenous compounds; includes nitrogen fixation, nitrification, denitrification, assimilatory/dissimilatory nitrate reduction and the interconversion of nitrogenous organic matter and ammonium.
organelle organization
A process that is carried out at the cellular level which results in the assembly, arrangement of constituent parts, or disassembly of an organelle within a cell. An organelle is an organized structure of distinctive morphology and function. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton. Excludes the plasma membrane.
microtubule-based process
Any cellular process that depends upon or alters the microtubule cytoskeleton, that part of the cytoskeleton comprising microtubules and their associated proteins.
microtubule-based movement
Movement of organelles, other microtubules and other particles along microtubules, mediated by motor proteins.
cell cycle
The progression of biochemical and morphological phases and events that occur in a cell during successive cell replication or nuclear replication events. Canonically, the cell cycle comprises the replication and segregation of genetic material followed by the division of the cell, but in endocycles or syncytial cells nuclear replication or nuclear division may not be followed by cell division.
mitosis
A cell cycle process comprising the steps by which the nucleus of a eukaryotic cell divides; the process involves condensation of chromosomal DNA into a highly compacted form. Canonically, mitosis produces two daughter nuclei whose chromosome complement is identical to that of the mother cell.
meiosis
A cell cycle process comprising the steps by which a cell progresses through the nuclear division phase of a meiotic cell cycle, the specialized nuclear and cell division in which a single diploid cell undergoes two nuclear divisions following a single round of DNA replication in order to produce four daughter cells that contain half the number of chromosomes as the diploid cell. Meiotic division occurs during the formation of gametes from diploid organisms and at the beginning of haplophase in those organisms that alternate between diploid and haploid generations.
meiosis I
A cell cycle process comprising the steps by which a cell progresses through the first phase of meiosis, in which cells divide and homologous chromosomes are paired and segregated from each other, producing two daughter cells.
synapsis
The cell cycle process whereby the side by side pairing and physical juxtaposition of homologous chromosomes is created at the metaphase plate.
gamete generation
The generation and maintenance of gametes in a multicellular organism. A gamete is a haploid reproductive cell.
spermatogenesis
The process of formation of spermatozoa, including spermatocytogenesis and spermiogenesis.
regulation of mitotic cell cycle
Any process that modulates the rate or extent of progress through the mitotic cell cycle.
biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
fertilization
The union of gametes of opposite sexes during the process of sexual reproduction to form a zygote. It involves the fusion of the gametic nuclei (karyogamy) and cytoplasm (plasmogamy).
cellular process
Any process that is carried out at the cellular level, but not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
cellular component organization
A process that is carried out at the cellular level which results in the assembly, arrangement of constituent parts, or disassembly of a cellular component.
RNA metabolic process
The cellular chemical reactions and pathways involving RNA, ribonucleic acid, one of the two main type of nucleic acid, consisting of a long, unbranched macromolecule formed from ribonucleotides joined in 3',5'-phosphodiester linkage.
sexual reproduction
The regular alternation, in the life cycle of haplontic, diplontic and diplohaplontic organisms, of meiosis and fertilization which provides for the production offspring. In diplontic organisms there is a life cycle in which the products of meiosis behave directly as gametes, fusing to form a zygote from which the diploid, or sexually reproductive polyploid, adult organism will develop. In diplohaplontic organisms a haploid phase (gametophyte) exists in the life cycle between meiosis and fertilization (e.g. higher plants, many algae and Fungi); the products of meiosis are spores that develop as haploid individuals from which haploid gametes develop to form a diploid zygote; diplohaplontic organisms show an alternation of haploid and diploid generations. In haplontic organisms meiosis occurs in the zygote, giving rise to four haploid cells (e.g. many algae and protozoa), only the zygote is diploid and this may form a resistant spore, tiding organisms over hard times.
cell cycle process
A cellular process that is involved in the progression of biochemical and morphological phases and events that occur in a cell during successive cell replication or nuclear replication events.
cell cycle phase
A cell cycle process comprising the steps by which a cell progresses through one of the biochemical and morphological phases and events that occur during successive cell replication or nuclear replication events.
reproductive process
A biological process that directly contributes to the process of producing new individuals by one or two organisms. The new individuals inherit some proportion of their genetic material from the parent or parents.
multicellular organismal process
Any biological process, occurring at the level of a multicellular organism, pertinent to its function.
multicellular organism reproduction
The biological process by which new individuals are produced by one or two multicellular organisms. The new individuals inherit some proportion of their genetic material from the parent or parents.
piRNA metabolic process
The chemical reactions and pathways involving piRNAs, Piwi-associated RNAs, a class of 24- to 30-nucleotide RNA derived from repeat or complex DNA sequence elements and processed by a Dicer-independent mechanism.
cellular nitrogen compound metabolic process
The chemical reactions and pathways involving various organic and inorganic nitrogenous compounds, as carried out by individual cells.
ncRNA metabolic process
The chemical reactions and pathways involving non-coding RNA transcripts (ncRNAs).
macromolecule metabolic process
The chemical reactions and pathways involving macromolecules, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass.
cellular metabolic process
The chemical reactions and pathways by which individual cells transform chemical substances.
primary metabolic process
The chemical reactions and pathways involving those compounds which are formed as a part of the normal anabolic and catabolic processes. These processes take place in most, if not all, cells of the organism.
cellular macromolecule metabolic process
The chemical reactions and pathways involving macromolecules, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass, as carried out by individual cells.
male gamete generation
Generation of the male gamete; specialised haploid cells produced by meiosis and along with a female gamete takes part in sexual reproduction.
organelle fission
The creation of two or more organelles by division of one organelle.
reproductive process in a multicellular organism
The process, occurring above the cellular level, that is pertinent to the reproductive function of a multicellular organism. This includes the integrated processes at the level of tissues and organs.
reproductive cellular process
A process, occurring at the cellular level, that is involved in the reproductive function of a multicellular or single-celled organism.
regulation of biological process
Any process that modulates the frequency, rate or extent of a biological process. Biological processes are regulated by many means; examples include the control of gene expression, protein modification or interaction with a protein or substrate molecule.
regulation of cellular process
Any process that modulates the frequency, rate or extent of a cellular process, any of those that are carried out at the cellular level, but are not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
cell division
The process resulting in the physical partitioning and separation of a cell into daughter cells.
meiotic cell cycle
Progression through the phases of the meiotic cell cycle, in which canonically a cell replicates to produce four offspring with half the chromosomal content of the progenitor cell.
M phase of meiotic cell cycle
A cell cycle process comprising the steps by which a cell progresses through M phase, the part of the meiotic cell cycle during which meiosis takes place.
biological regulation
Any process that modulates the frequency, rate or extent of any biological process, quality or function.
chromosome organization involved in meiosis
A process of chromosome organization that is involved in meiosis.
nucleic acid metabolic process
Any cellular metabolic process involving nucleic acids.
all
NA
reproductive process
A biological process that directly contributes to the process of producing new individuals by one or two organisms. The new individuals inherit some proportion of their genetic material from the parent or parents.
cellular metabolic process
The chemical reactions and pathways by which individual cells transform chemical substances.
organelle organization
A process that is carried out at the cellular level which results in the assembly, arrangement of constituent parts, or disassembly of an organelle within a cell. An organelle is an organized structure of distinctive morphology and function. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton. Excludes the plasma membrane.
reproductive cellular process
A process, occurring at the cellular level, that is involved in the reproductive function of a multicellular or single-celled organism.
multicellular organism reproduction
The biological process by which new individuals are produced by one or two multicellular organisms. The new individuals inherit some proportion of their genetic material from the parent or parents.
reproductive process in a multicellular organism
The process, occurring above the cellular level, that is pertinent to the reproductive function of a multicellular organism. This includes the integrated processes at the level of tissues and organs.
regulation of cellular process
Any process that modulates the frequency, rate or extent of a cellular process, any of those that are carried out at the cellular level, but are not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
regulation of biological process
Any process that modulates the frequency, rate or extent of a biological process. Biological processes are regulated by many means; examples include the control of gene expression, protein modification or interaction with a protein or substrate molecule.
fertilization
The union of gametes of opposite sexes during the process of sexual reproduction to form a zygote. It involves the fusion of the gametic nuclei (karyogamy) and cytoplasm (plasmogamy).
reproductive process in a multicellular organism
The process, occurring above the cellular level, that is pertinent to the reproductive function of a multicellular organism. This includes the integrated processes at the level of tissues and organs.
cellular nitrogen compound metabolic process
The chemical reactions and pathways involving various organic and inorganic nitrogenous compounds, as carried out by individual cells.
cellular macromolecule metabolic process
The chemical reactions and pathways involving macromolecules, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass, as carried out by individual cells.
cell cycle process
A cellular process that is involved in the progression of biochemical and morphological phases and events that occur in a cell during successive cell replication or nuclear replication events.
regulation of cell cycle
Any process that modulates the rate or extent of progression through the cell cycle.
gamete generation
The generation and maintenance of gametes in a multicellular organism. A gamete is a haploid reproductive cell.
nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
Any cellular metabolic process involving nucleobases, nucleosides, nucleotides and nucleic acids.
chromosome organization involved in meiosis
A process of chromosome organization that is involved in meiosis.
microtubule cytoskeleton organization
A process that is carried out at the cellular level which results in the assembly, arrangement of constituent parts, or disassembly of cytoskeletal structures comprising microtubules and their associated proteins.
regulation of mitotic cell cycle
Any process that modulates the rate or extent of progress through the mitotic cell cycle.
RNA metabolic process
The cellular chemical reactions and pathways involving RNA, ribonucleic acid, one of the two main type of nucleic acid, consisting of a long, unbranched macromolecule formed from ribonucleotides joined in 3',5'-phosphodiester linkage.
mitosis
A cell cycle process comprising the steps by which the nucleus of a eukaryotic cell divides; the process involves condensation of chromosomal DNA into a highly compacted form. Canonically, mitosis produces two daughter nuclei whose chromosome complement is identical to that of the mother cell.
mitosis
A cell cycle process comprising the steps by which the nucleus of a eukaryotic cell divides; the process involves condensation of chromosomal DNA into a highly compacted form. Canonically, mitosis produces two daughter nuclei whose chromosome complement is identical to that of the mother cell.
meiosis
A cell cycle process comprising the steps by which a cell progresses through the nuclear division phase of a meiotic cell cycle, the specialized nuclear and cell division in which a single diploid cell undergoes two nuclear divisions following a single round of DNA replication in order to produce four daughter cells that contain half the number of chromosomes as the diploid cell. Meiotic division occurs during the formation of gametes from diploid organisms and at the beginning of haplophase in those organisms that alternate between diploid and haploid generations.
M phase of mitotic cell cycle
A cell cycle process comprising the steps by which a cell progresses through M phase, the part of the mitotic cell cycle during which mitosis takes place.
M phase of meiotic cell cycle
A cell cycle process comprising the steps by which a cell progresses through M phase, the part of the meiotic cell cycle during which meiosis takes place.
meiosis I
A cell cycle process comprising the steps by which a cell progresses through the first phase of meiosis, in which cells divide and homologous chromosomes are paired and segregated from each other, producing two daughter cells.
chromosome organization involved in meiosis
A process of chromosome organization that is involved in meiosis.
synapsis
The cell cycle process whereby the side by side pairing and physical juxtaposition of homologous chromosomes is created at the metaphase plate.

nuclear chromosome
A chromosome found in the nucleus of a eukaryotic cell.
condensed chromosome
A highly compacted molecule of DNA and associated proteins resulting in a cytologically distinct structure.
intracellular
The living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
chromosome, centromeric region
The region of a chromosome that includes the centromeric DNA and associated proteins. In monocentric chromosomes, this region corresponds to a single area of the chromosome, whereas in holocentric chromosomes, it is evenly distributed along the chromosome.
kinetochore
A multisubunit complex that is located at the centromeric region of DNA and provides an attachment point for the spindle microtubules.
condensed chromosome kinetochore
A multisubunit complex that is located at the centromeric region of a condensed chromosome and provides an attachment point for the spindle microtubules.
condensed chromosome, centromeric region
The region of a condensed chromosome that includes the centromere and associated proteins, including the kinetochore. In monocentric chromosomes, this region corresponds to a single area of the chromosome, whereas in holocentric chromosomes, it is evenly distributed along the chromosome.
ribonucleoprotein complex
A macromolecular complex containing both protein and RNA molecules.
axoneme
The bundle of microtubules and associated proteins that forms the core of cilia and flagella in eukaryotic cells and is responsible for their movements.
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
cell
The basic structural and functional unit of all organisms. Includes the plasma membrane and any external encapsulating structures such as the cell wall and cell envelope.
nucleus
A membrane-bounded organelle of eukaryotic cells in which chromosomes are housed and replicated. In most cells, the nucleus contains all of the cell's chromosomes except the organellar chromosomes, and is the site of RNA synthesis and processing. In some species, or in specialized cell types, RNA metabolism or DNA replication may be absent.
cytoplasm
All of the contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures.
chromosome
A structure composed of a very long molecule of DNA and associated proteins (e.g. histones) that carries hereditary information.
spindle
The array of microtubules and associated molecules that forms between opposite poles of a eukaryotic cell during mitosis or meiosis and serves to move the duplicated chromosomes apart.
cytoskeleton
Any of the various filamentous elements that form the internal framework of cells, and typically remain after treatment of the cells with mild detergent to remove membrane constituents and soluble components of the cytoplasm. The term embraces intermediate filaments, microfilaments, microtubules, the microtrabecular lattice, and other structures characterized by a polymeric filamentous nature and long-range order within the cell. The various elements of the cytoskeleton not only serve in the maintenance of cellular shape but also have roles in other cellular functions, including cellular movement, cell division, endocytosis, and movement of organelles.
microtubule
Any of the long, generally straight, hollow tubes of internal diameter 12-15 nm and external diameter 24 nm found in a wide variety of eukaryotic cells; each consists (usually) of 13 protofilaments of polymeric tubulin, staggered in such a manner that the tubulin monomers are arranged in a helical pattern on the microtubular surface, and with the alpha/beta axes of the tubulin subunits parallel to the long axis of the tubule; exist in equilibrium with pool of tubulin monomers and can be rapidly assembled or disassembled in response to physiological stimuli; concerned with force generation, e.g. in the spindle.
cilium
A specialized eukaryotic organelle that consists of a filiform extrusion of the cell surface. Each cilium is bounded by an extrusion of the cytoplasmic membrane, and contains a regular longitudinal array of microtubules, anchored basally in a centriole.
microtubule cytoskeleton
The part of the cytoskeleton (the internal framework of a cell) composed of microtubules and associated proteins.
macromolecular complex
A stable assembly of two or more macromolecules, i.e. proteins, nucleic acids, carbohydrates or lipids, in which the constituent parts function together.
chromatoid body
A ribonucleoprotein complex found in the cytoplasm of male germ cells, composed of exceedingly thin filaments that are consolidated into a compact mass or into dense strands of varying thickness that branch to form an irregular network. Contains mRNAs, miRNAs, and protein components involved in miRNA processing (such as Argonaute proteins and the endonuclease Dicer) and in RNA decay (such as the decapping enzyme DCP1a and GW182).
cell projection
A prolongation or process extending from a cell, e.g. a flagellum or axon.
P granule
A small cytoplasmic, non-membranous RNA/protein complex aggregates in the primordial germ cells of many higher eukaryotes.
organelle
Organized structure of distinctive morphology and function. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton, and prokaryotic structures such as anammoxosomes and pirellulosomes. Excludes the plasma membrane.
membrane-bounded organelle
Organized structure of distinctive morphology and function, bounded by a single or double lipid bilayer membrane. Includes the nucleus, mitochondria, plastids, vacuoles, and vesicles. Excludes the plasma membrane.
non-membrane-bounded organelle
Organized structure of distinctive morphology and function, not bounded by a lipid bilayer membrane. Includes ribosomes, the cytoskeleton and chromosomes.
intracellular organelle
Organized structure of distinctive morphology and function, occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton. Excludes the plasma membrane.
intracellular membrane-bounded organelle
Organized structure of distinctive morphology and function, bounded by a single or double lipid bilayer membrane and occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, and vesicles. Excludes the plasma membrane.
intracellular non-membrane-bounded organelle
Organized structure of distinctive morphology and function, not bounded by a lipid bilayer membrane and occurring within the cell. Includes ribosomes, the cytoskeleton and chromosomes.
protein complex
Any macromolecular complex composed of two or more polypeptide subunits, which may or may not be identical. Protein complexes may have other associated non-protein prosthetic groups, such as nucleotides, metal ions or other small molecules.
organelle part
Any constituent part of an organelle, an organized structure of distinctive morphology and function. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton, but excludes the plasma membrane.
intracellular part
Any constituent part of the living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
chromosomal part
Any constituent part of a chromosome, a structure composed of a very long molecule of DNA and associated proteins (e.g. histones) that carries hereditary information.
nuclear part
Any constituent part of the nucleus, a membrane-bounded organelle of eukaryotic cells in which chromosomes are housed and replicated.
cytoskeletal part
Any constituent part of the cytoskeleton, a cellular scaffolding or skeleton that maintains cell shape, enables some cell motion (using structures such as flagella and cilia), and plays important roles in both intra-cellular transport (e.g. the movement of vesicles and organelles) and cellular division. Includes constituent parts of intermediate filaments, microfilaments, microtubules, and the microtrabecular lattice.
cilium part
Any constituent part of a cilium, a specialized eukaryotic organelle that consists of a filiform extrusion of the cell surface. Each cilium is bounded by an extrusion of the cytoplasmic membrane, and contains a regular longitudinal array of microtubules, anchored basally in a centriole.
cytoplasmic part
Any constituent part of the cytoplasm, all of the contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures.
intracellular organelle part
A constituent part of an intracellular organelle, an organized structure of distinctive morphology and function, occurring within the cell. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton but excludes the plasma membrane.
cell projection part
Any constituent part of a cell projection, a prolongation or process extending from a cell, e.g. a flagellum or axon.
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
pole plasm
Differentiated cytoplasm associated with a pole (animal, vegetal, anterior, or posterior) of an oocyte, egg or early embryo.
germ plasm
Differentiated cytoplasm associated with a pole of an oocyte, egg or early embryo that will be inherited by the cells that will give rise to the germ line.
all
NA
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
organelle part
Any constituent part of an organelle, an organized structure of distinctive morphology and function. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton, but excludes the plasma membrane.
intracellular membrane-bounded organelle
Organized structure of distinctive morphology and function, bounded by a single or double lipid bilayer membrane and occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, and vesicles. Excludes the plasma membrane.
intracellular non-membrane-bounded organelle
Organized structure of distinctive morphology and function, not bounded by a lipid bilayer membrane and occurring within the cell. Includes ribosomes, the cytoskeleton and chromosomes.
intracellular organelle part
A constituent part of an intracellular organelle, an organized structure of distinctive morphology and function, occurring within the cell. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton but excludes the plasma membrane.
intracellular part
Any constituent part of the living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
cell projection part
Any constituent part of a cell projection, a prolongation or process extending from a cell, e.g. a flagellum or axon.
ribonucleoprotein complex
A macromolecular complex containing both protein and RNA molecules.
axoneme
The bundle of microtubules and associated proteins that forms the core of cilia and flagella in eukaryotic cells and is responsible for their movements.
intracellular organelle
Organized structure of distinctive morphology and function, occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton. Excludes the plasma membrane.
intracellular organelle part
A constituent part of an intracellular organelle, an organized structure of distinctive morphology and function, occurring within the cell. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton but excludes the plasma membrane.
axoneme
The bundle of microtubules and associated proteins that forms the core of cilia and flagella in eukaryotic cells and is responsible for their movements.
cilium part
Any constituent part of a cilium, a specialized eukaryotic organelle that consists of a filiform extrusion of the cell surface. Each cilium is bounded by an extrusion of the cytoplasmic membrane, and contains a regular longitudinal array of microtubules, anchored basally in a centriole.
axoneme
The bundle of microtubules and associated proteins that forms the core of cilia and flagella in eukaryotic cells and is responsible for their movements.
kinetochore
A multisubunit complex that is located at the centromeric region of DNA and provides an attachment point for the spindle microtubules.
kinetochore
A multisubunit complex that is located at the centromeric region of DNA and provides an attachment point for the spindle microtubules.
spindle
The array of microtubules and associated molecules that forms between opposite poles of a eukaryotic cell during mitosis or meiosis and serves to move the duplicated chromosomes apart.
microtubule
Any of the long, generally straight, hollow tubes of internal diameter 12-15 nm and external diameter 24 nm found in a wide variety of eukaryotic cells; each consists (usually) of 13 protofilaments of polymeric tubulin, staggered in such a manner that the tubulin monomers are arranged in a helical pattern on the microtubular surface, and with the alpha/beta axes of the tubulin subunits parallel to the long axis of the tubule; exist in equilibrium with pool of tubulin monomers and can be rapidly assembled or disassembled in response to physiological stimuli; concerned with force generation, e.g. in the spindle.
cilium part
Any constituent part of a cilium, a specialized eukaryotic organelle that consists of a filiform extrusion of the cell surface. Each cilium is bounded by an extrusion of the cytoplasmic membrane, and contains a regular longitudinal array of microtubules, anchored basally in a centriole.
cytoplasmic part
Any constituent part of the cytoplasm, all of the contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures.
P granule
A small cytoplasmic, non-membranous RNA/protein complex aggregates in the primordial germ cells of many higher eukaryotes.
nuclear part
Any constituent part of the nucleus, a membrane-bounded organelle of eukaryotic cells in which chromosomes are housed and replicated.
nuclear chromosome
A chromosome found in the nucleus of a eukaryotic cell.
chromosomal part
Any constituent part of a chromosome, a structure composed of a very long molecule of DNA and associated proteins (e.g. histones) that carries hereditary information.
cytoskeletal part
Any constituent part of the cytoskeleton, a cellular scaffolding or skeleton that maintains cell shape, enables some cell motion (using structures such as flagella and cilia), and plays important roles in both intra-cellular transport (e.g. the movement of vesicles and organelles) and cellular division. Includes constituent parts of intermediate filaments, microfilaments, microtubules, and the microtrabecular lattice.
kinetochore
A multisubunit complex that is located at the centromeric region of DNA and provides an attachment point for the spindle microtubules.
condensed chromosome, centromeric region
The region of a condensed chromosome that includes the centromere and associated proteins, including the kinetochore. In monocentric chromosomes, this region corresponds to a single area of the chromosome, whereas in holocentric chromosomes, it is evenly distributed along the chromosome.
spindle
The array of microtubules and associated molecules that forms between opposite poles of a eukaryotic cell during mitosis or meiosis and serves to move the duplicated chromosomes apart.
microtubule
Any of the long, generally straight, hollow tubes of internal diameter 12-15 nm and external diameter 24 nm found in a wide variety of eukaryotic cells; each consists (usually) of 13 protofilaments of polymeric tubulin, staggered in such a manner that the tubulin monomers are arranged in a helical pattern on the microtubular surface, and with the alpha/beta axes of the tubulin subunits parallel to the long axis of the tubule; exist in equilibrium with pool of tubulin monomers and can be rapidly assembled or disassembled in response to physiological stimuli; concerned with force generation, e.g. in the spindle.
condensed chromosome kinetochore
A multisubunit complex that is located at the centromeric region of a condensed chromosome and provides an attachment point for the spindle microtubules.
P granule
A small cytoplasmic, non-membranous RNA/protein complex aggregates in the primordial germ cells of many higher eukaryotes.

molecular_function
Elemental activities, such as catalysis or binding, describing the actions of a gene product at the molecular level. A given gene product may exhibit one or more molecular functions.
nucleic acid binding
Interacting selectively and non-covalently with any nucleic acid.
motor activity
Catalysis of movement along a polymeric molecule such as a microfilament or microtubule, coupled to the hydrolysis of a nucleoside triphosphate.
microtubule motor activity
Catalysis of movement along a microtubule, coupled to the hydrolysis of a nucleoside triphosphate (usually ATP).
catalytic activity
Catalysis of a biochemical reaction at physiological temperatures. In biologically catalyzed reactions, the reactants are known as substrates, and the catalysts are naturally occurring macromolecular substances known as enzymes. Enzymes possess specific binding sites for substrates, and are usually composed wholly or largely of protein, but RNA that has catalytic activity (ribozyme) is often also regarded as enzymatic.
nucleoside-triphosphatase activity
Catalysis of the reaction: a nucleoside triphosphate + H2O = nucleoside diphosphate + phosphate.
binding
The selective, non-covalent, often stoichiometric, interaction of a molecule with one or more specific sites on another molecule.
pyrophosphatase activity
Catalysis of the hydrolysis of a pyrophosphate bond between two phosphate groups, leaving one phosphate on each of the two fragments.
hydrolase activity
Catalysis of the hydrolysis of various bonds, e.g. C-O, C-N, C-C, phosphoric anhydride bonds, etc. Hydrolase is the systematic name for any enzyme of EC class 3.
hydrolase activity, acting on acid anhydrides
Catalysis of the hydrolysis of any acid anhydride.
hydrolase activity, acting on acid anhydrides, in phosphorus-containing anhydrides
Catalysis of the hydrolysis of any acid anhydride which contains phosphorus.
all
NA
ADAD1adenosine deaminase domain containing 1 (testis-specific) (ENSG00000164113), score: 0.53 ADAM10ADAM metallopeptidase domain 10 (ENSG00000137845), score: -0.51 ADPRHL2ADP-ribosylhydrolase like 2 (ENSG00000116863), score: -0.58 AGBL3ATP/GTP binding protein-like 3 (ENSG00000146856), score: 0.67 AGFG1ArfGAP with FG repeats 1 (ENSG00000173744), score: 0.52 AIG1androgen-induced 1 (ENSG00000146416), score: -0.51 AK7adenylate kinase 7 (ENSG00000140057), score: 0.62 AKIRIN2akirin 2 (ENSG00000135334), score: 0.62 ANAPC10anaphase promoting complex subunit 10 (ENSG00000164162), score: 0.6 ANGEL1angel homolog 1 (Drosophila) (ENSG00000013523), score: 0.53 ANKFN1ankyrin-repeat and fibronectin type III domain containing 1 (ENSG00000153930), score: 0.66 ANKRD5ankyrin repeat domain 5 (ENSG00000132623), score: 0.66 ANUBL1AN1, ubiquitin-like, homolog (Xenopus laevis) (ENSG00000172671), score: 0.51 ANXA6annexin A6 (ENSG00000197043), score: -0.55 ANXA7annexin A7 (ENSG00000138279), score: -0.65 APBB2amyloid beta (A4) precursor protein-binding, family B, member 2 (ENSG00000163697), score: -0.58 APPamyloid beta (A4) precursor protein (ENSG00000142192), score: -0.51 APTXaprataxin (ENSG00000137074), score: 0.53 ARID2AT rich interactive domain 2 (ARID, RFX-like) (ENSG00000189079), score: 0.58 ARID3BAT rich interactive domain 3B (BRIGHT-like) (ENSG00000179361), score: 0.57 ARL13BADP-ribosylation factor-like 13B (ENSG00000169379), score: 0.73 ARL9ADP-ribosylation factor-like 9 (ENSG00000196503), score: 0.5 ARMC3armadillo repeat containing 3 (ENSG00000165309), score: 0.58 ARMC4armadillo repeat containing 4 (ENSG00000169126), score: 0.5 ASPMasp (abnormal spindle) homolog, microcephaly associated (Drosophila) (ENSG00000066279), score: 0.72 AURKAaurora kinase A (ENSG00000087586), score: 0.6 BAZ1Abromodomain adjacent to zinc finger domain, 1A (ENSG00000198604), score: 0.5 BBS7Bardet-Biedl syndrome 7 (ENSG00000138686), score: 0.53 BHLHE40basic helix-loop-helix family, member e40 (ENSG00000134107), score: -0.53 BLMBloom syndrome, RecQ helicase-like (ENSG00000197299), score: 0.51 BOLLbol, boule-like (Drosophila) (ENSG00000152430), score: 0.55 BRAPBRCA1 associated protein (ENSG00000089234), score: 0.52 BRCA2breast cancer 2, early onset (ENSG00000139618), score: 0.7 BUB1budding uninhibited by benzimidazoles 1 homolog (yeast) (ENSG00000169679), score: 0.52 BUB1Bbudding uninhibited by benzimidazoles 1 homolog beta (yeast) (ENSG00000156970), score: 0.54 C10orf46chromosome 10 open reading frame 46 (ENSG00000151893), score: 0.51 C11orf70chromosome 11 open reading frame 70 (ENSG00000137691), score: 0.54 C11orf82chromosome 11 open reading frame 82 (ENSG00000165490), score: 0.62 C12orf48chromosome 12 open reading frame 48 (ENSG00000185480), score: 0.54 C12orf50chromosome 12 open reading frame 50 (ENSG00000165805), score: 0.59 C12orf63chromosome 12 open reading frame 63 (ENSG00000188596), score: 0.65 C13orf34chromosome 13 open reading frame 34 (ENSG00000136122), score: 0.69 C14orf159chromosome 14 open reading frame 159 (ENSG00000133943), score: -0.53 C14orf166Bchromosome 14 open reading frame 166B (ENSG00000100565), score: 0.64 C14orf38chromosome 14 open reading frame 38 (ENSG00000151838), score: 0.62 C14orf39chromosome 14 open reading frame 39 (ENSG00000179008), score: 0.51 C14orf49chromosome 14 open reading frame 49 (ENSG00000176438), score: 0.56 C14orf50chromosome 14 open reading frame 50 (ENSG00000165807), score: 0.65 C15orf26chromosome 15 open reading frame 26 (ENSG00000156206), score: 0.69 C17orf104chromosome 17 open reading frame 104 (ENSG00000180336), score: 0.58 C19orf45chromosome 19 open reading frame 45 (ENSG00000198723), score: 0.69 C1orf100chromosome 1 open reading frame 100 (ENSG00000173728), score: 0.64 C1orf111chromosome 1 open reading frame 111 (ENSG00000171722), score: 0.65 C1orf158chromosome 1 open reading frame 158 (ENSG00000157330), score: 0.6 C1orf228chromosome 1 open reading frame 228 (ENSG00000198520), score: 0.6 C1orf88chromosome 1 open reading frame 88 (ENSG00000173947), score: 0.52 C1orf9chromosome 1 open reading frame 9 (ENSG00000094975), score: 0.51 C20orf85chromosome 20 open reading frame 85 (ENSG00000124237), score: 0.66 C2orf65chromosome 2 open reading frame 65 (ENSG00000159374), score: 0.78 C2orf71chromosome 2 open reading frame 71 (ENSG00000179270), score: 0.71 C3orf38chromosome 3 open reading frame 38 (ENSG00000179021), score: 0.55 C4orf47chromosome 4 open reading frame 47 (ENSG00000205129), score: 0.54 C5orf28chromosome 5 open reading frame 28 (ENSG00000151881), score: 0.51 C6orf150chromosome 6 open reading frame 150 (ENSG00000164430), score: 0.67 C6orf204chromosome 6 open reading frame 204 (ENSG00000111860), score: 0.55 C6orf64chromosome 6 open reading frame 64 (ENSG00000112167), score: 0.56 C7orf31chromosome 7 open reading frame 31 (ENSG00000153790), score: 0.55 C7orf45chromosome 7 open reading frame 45 (ENSG00000165120), score: 0.75 C7orf57chromosome 7 open reading frame 57 (ENSG00000164746), score: 0.57 C7orf62chromosome 7 open reading frame 62 (ENSG00000164645), score: 0.58 C9orf9chromosome 9 open reading frame 9 (ENSG00000165698), score: 0.51 C9orf96chromosome 9 open reading frame 96 (ENSG00000198870), score: 0.53 C9orf98chromosome 9 open reading frame 98 (ENSG00000165695), score: 0.56 CACNA1Scalcium channel, voltage-dependent, L type, alpha 1S subunit (ENSG00000081248), score: 0.51 CALR3calreticulin 3 (ENSG00000141979), score: 0.54 CASC1cancer susceptibility candidate 1 (ENSG00000118307), score: 0.5 CCDC108coiled-coil domain containing 108 (ENSG00000181378), score: 0.67 CCDC146coiled-coil domain containing 146 (ENSG00000135205), score: 0.53 CCDC27coiled-coil domain containing 27 (ENSG00000162592), score: 0.67 CCDC30coiled-coil domain containing 30 (ENSG00000186409), score: 0.61 CCDC37coiled-coil domain containing 37 (ENSG00000163885), score: 0.57 CCDC40coiled-coil domain containing 40 (ENSG00000141519), score: 0.61 CCDC41coiled-coil domain containing 41 (ENSG00000173588), score: 0.64 CCDC47coiled-coil domain containing 47 (ENSG00000108588), score: -0.53 CCDC63coiled-coil domain containing 63 (ENSG00000173093), score: 0.72 CCDC67coiled-coil domain containing 67 (ENSG00000165325), score: 0.7 CCDC83coiled-coil domain containing 83 (ENSG00000150676), score: 0.56 CCDC90Acoiled-coil domain containing 90A (ENSG00000050393), score: -0.51 CCNA1cyclin A1 (ENSG00000133101), score: 0.55 CCR4chemokine (C-C motif) receptor 4 (ENSG00000183813), score: 0.64 CCT4chaperonin containing TCP1, subunit 4 (delta) (ENSG00000115484), score: 0.57 CDKN3cyclin-dependent kinase inhibitor 3 (ENSG00000100526), score: 0.58 CENPIcentromere protein I (ENSG00000102384), score: 0.56 CENPKcentromere protein K (ENSG00000123219), score: 0.59 CENPNcentromere protein N (ENSG00000166451), score: 0.53 CENPQcentromere protein Q (ENSG00000031691), score: 0.52 CGRRF1cell growth regulator with ring finger domain 1 (ENSG00000100532), score: 0.5 CHRNA9cholinergic receptor, nicotinic, alpha 9 (ENSG00000174343), score: 0.83 CNOT4CCR4-NOT transcription complex, subunit 4 (ENSG00000080802), score: 0.82 CRYBA1crystallin, beta A1 (ENSG00000108255), score: 0.71 CSNK2A1casein kinase 2, alpha 1 polypeptide (ENSG00000101266), score: -0.55 CSTF1cleavage stimulation factor, 3' pre-RNA, subunit 1, 50kDa (ENSG00000101138), score: 0.5 CXorf22chromosome X open reading frame 22 (ENSG00000165164), score: 0.74 CXorf30chromosome X open reading frame 30 (ENSG00000205081), score: 0.61 DDX4DEAD (Asp-Glu-Ala-Asp) box polypeptide 4 (ENSG00000152670), score: 0.53 DIS3DIS3 mitotic control homolog (S. cerevisiae) (ENSG00000083520), score: 0.58 DMRT3doublesex and mab-3 related transcription factor 3 (ENSG00000064218), score: 0.64 DNAH8dynein, axonemal, heavy chain 8 (ENSG00000124721), score: 0.76 DNAI1dynein, axonemal, intermediate chain 1 (ENSG00000122735), score: 0.53 DNAI2dynein, axonemal, intermediate chain 2 (ENSG00000171595), score: 0.63 DYDC1DPY30 domain containing 1 (ENSG00000170788), score: 0.55 DYMdymeclin (ENSG00000141627), score: 0.52 E2F8E2F transcription factor 8 (ENSG00000129173), score: 0.68 EFCAB5EF-hand calcium binding domain 5 (ENSG00000176927), score: 0.67 EFHC1EF-hand domain (C-terminal) containing 1 (ENSG00000096093), score: 0.58 ENO4enolase family member 4 (ENSG00000188316), score: 0.54 EPYCepiphycan (ENSG00000083782), score: 0.57 ESAMendothelial cell adhesion molecule (ENSG00000149564), score: -0.51 EXO1exonuclease 1 (ENSG00000174371), score: 0.52 EYA4eyes absent homolog 4 (Drosophila) (ENSG00000112319), score: 0.58 FAM124Bfamily with sequence similarity 124B (ENSG00000124019), score: 0.55 FAM194Afamily with sequence similarity 194, member A (ENSG00000163645), score: 0.51 FAM73Bfamily with sequence similarity 73, member B (ENSG00000148343), score: -0.53 FAM76Afamily with sequence similarity 76, member A (ENSG00000009780), score: 0.52 FBXO30F-box protein 30 (ENSG00000118496), score: 0.61 FBXO43F-box protein 43 (ENSG00000156509), score: 0.62 FBXO47F-box protein 47 (ENSG00000204952), score: 0.72 FER1L6fer-1-like 6 (C. elegans) (ENSG00000214814), score: 0.58 FHDC1FH2 domain containing 1 (ENSG00000137460), score: 0.58 FIGNL1fidgetin-like 1 (ENSG00000132436), score: 0.66 FNDC3Afibronectin type III domain containing 3A (ENSG00000102531), score: 0.61 FNDC7fibronectin type III domain containing 7 (ENSG00000143107), score: 0.5 FYTTD1forty-two-three domain containing 1 (ENSG00000122068), score: 0.59 GCNT3glucosaminyl (N-acetyl) transferase 3, mucin type (ENSG00000140297), score: 0.54 GEMIN5gem (nuclear organelle) associated protein 5 (ENSG00000082516), score: 0.59 GKAP1G kinase anchoring protein 1 (ENSG00000165113), score: 0.57 GLG1golgi glycoprotein 1 (ENSG00000090863), score: -0.52 GLT8D1glycosyltransferase 8 domain containing 1 (ENSG00000016864), score: 0.65 GMPSguanine monphosphate synthetase (ENSG00000163655), score: 0.56 GPATCH2G patch domain containing 2 (ENSG00000092978), score: 0.65 GPR160G protein-coupled receptor 160 (ENSG00000173890), score: 0.53 GRB2growth factor receptor-bound protein 2 (ENSG00000177885), score: -0.57 GRHL1grainyhead-like 1 (Drosophila) (ENSG00000134317), score: 0.55 HELLShelicase, lymphoid-specific (ENSG00000119969), score: 0.56 HERC4hect domain and RLD 4 (ENSG00000148634), score: 0.52 HMOX2heme oxygenase (decycling) 2 (ENSG00000103415), score: 0.56 HORMAD1HORMA domain containing 1 (ENSG00000143452), score: 0.57 IFT172intraflagellar transport 172 homolog (Chlamydomonas) (ENSG00000138002), score: 0.53 IFT46intraflagellar transport 46 homolog (Chlamydomonas) (ENSG00000118096), score: 0.53 IFT74intraflagellar transport 74 homolog (Chlamydomonas) (ENSG00000096872), score: 0.58 IGF2BP1insulin-like growth factor 2 mRNA binding protein 1 (ENSG00000159217), score: 0.59 IMPG2interphotoreceptor matrix proteoglycan 2 (ENSG00000081148), score: 0.5 IQCHIQ motif containing H (ENSG00000103599), score: 0.67 IQUBIQ motif and ubiquitin domain containing (ENSG00000164675), score: 0.67 KATNAL2katanin p60 subunit A-like 2 (ENSG00000167216), score: 0.59 KCTD21potassium channel tetramerisation domain containing 21 (ENSG00000188997), score: -0.54 KIAA0586KIAA0586 (ENSG00000100578), score: 0.63 KIF11kinesin family member 11 (ENSG00000138160), score: 0.51 KIF14kinesin family member 14 (ENSG00000118193), score: 0.66 KIF18Akinesin family member 18A (ENSG00000121621), score: 0.63 KIF18Bkinesin family member 18B (ENSG00000186185), score: 0.95 KIF24kinesin family member 24 (ENSG00000186638), score: 0.61 KIF27kinesin family member 27 (ENSG00000165115), score: 0.6 KIF2Bkinesin family member 2B (ENSG00000141200), score: 0.64 KLHL10kelch-like 10 (Drosophila) (ENSG00000161594), score: 0.66 KNTC1kinetochore associated 1 (ENSG00000184445), score: 0.52 KPNA6karyopherin alpha 6 (importin alpha 7) (ENSG00000025800), score: 0.81 LASS3LAG1 homolog, ceramide synthase 3 (ENSG00000154227), score: 0.61 LGSNlengsin, lens protein with glutamine synthetase domain (ENSG00000146166), score: 0.59 LIN28Blin-28 homolog B (C. elegans) (ENSG00000187772), score: 0.57 LPXNleupaxin (ENSG00000110031), score: 0.53 LRGUKleucine-rich repeats and guanylate kinase domain containing (ENSG00000155530), score: 0.58 LRRC18leucine rich repeat containing 18 (ENSG00000165383), score: 0.55 LRRC43leucine rich repeat containing 43 (ENSG00000158113), score: 0.66 LRRC48leucine rich repeat containing 48 (ENSG00000171962), score: 0.6 LRRC50leucine rich repeat containing 50 (ENSG00000154099), score: 0.51 LRRC67leucine rich repeat containing 67 (ENSG00000178125), score: 0.76 LRRC8Aleucine rich repeat containing 8 family, member A (ENSG00000136802), score: -0.6 LRRIQ4leucine-rich repeats and IQ motif containing 4 (ENSG00000188306), score: 0.68 M6PRmannose-6-phosphate receptor (cation dependent) (ENSG00000003056), score: -0.53 MAELmaelstrom homolog (Drosophila) (ENSG00000143194), score: 0.54 MAKmale germ cell-associated kinase (ENSG00000111837), score: 0.53 MAS1MAS1 oncogene (ENSG00000130368), score: 0.5 MAT2Bmethionine adenosyltransferase II, beta (ENSG00000038274), score: -0.52 MCM9minichromosome maintenance complex component 9 (ENSG00000111877), score: 0.51 MDH1Bmalate dehydrogenase 1B, NAD (soluble) (ENSG00000138400), score: 0.59 MELKmaternal embryonic leucine zipper kinase (ENSG00000165304), score: 0.5 MLXMAX-like protein X (ENSG00000108788), score: -0.6 MORN3MORN repeat containing 3 (ENSG00000139714), score: 0.55 MSL1male-specific lethal 1 homolog (Drosophila) (ENSG00000188895), score: 0.52 MYBL1v-myb myeloblastosis viral oncogene homolog (avian)-like 1 (ENSG00000185697), score: 0.65 MYF5myogenic factor 5 (ENSG00000111049), score: 0.81 MYF6myogenic factor 6 (herculin) (ENSG00000111046), score: 1 NCAPGnon-SMC condensin I complex, subunit G (ENSG00000109805), score: 0.62 NCBP1nuclear cap binding protein subunit 1, 80kDa (ENSG00000136937), score: 0.72 NEK2NIMA (never in mitosis gene a)-related kinase 2 (ENSG00000117650), score: 0.62 NFIBnuclear factor I/B (ENSG00000147862), score: -0.55 NMBRneuromedin B receptor (ENSG00000135577), score: 0.87 NRD1nardilysin (N-arginine dibasic convertase) (ENSG00000078618), score: 0.54 NUF2NUF2, NDC80 kinetochore complex component, homolog (S. cerevisiae) (ENSG00000143228), score: 0.58 NUP205nucleoporin 205kDa (ENSG00000155561), score: 0.71 ORC1Lorigin recognition complex, subunit 1-like (S. cerevisiae) (ENSG00000085840), score: 0.64 PACRGPARK2 co-regulated (ENSG00000112530), score: 0.5 PAK1IP1PAK1 interacting protein 1 (ENSG00000111845), score: 0.53 PASKPAS domain containing serine/threonine kinase (ENSG00000115687), score: 0.5 PCGF2polycomb group ring finger 2 (ENSG00000056661), score: -0.51 PDCD4programmed cell death 4 (neoplastic transformation inhibitor) (ENSG00000150593), score: 0.54 PELI2pellino homolog 2 (Drosophila) (ENSG00000139946), score: 0.57 PFN4profilin family, member 4 (ENSG00000176732), score: 0.51 PHF13PHD finger protein 13 (ENSG00000116273), score: 0.5 PIAS1protein inhibitor of activated STAT, 1 (ENSG00000033800), score: 0.52 PIWIL1piwi-like 1 (Drosophila) (ENSG00000125207), score: 0.56 PKD2L1polycystic kidney disease 2-like 1 (ENSG00000107593), score: 0.5 PKD2L2polycystic kidney disease 2-like 2 (ENSG00000078795), score: 0.5 POC5POC5 centriolar protein homolog (Chlamydomonas) (ENSG00000152359), score: 0.51 POLD3polymerase (DNA-directed), delta 3, accessory subunit (ENSG00000077514), score: 0.6 POU2F3POU class 2 homeobox 3 (ENSG00000137709), score: 0.63 PRDM4PR domain containing 4 (ENSG00000110851), score: 0.64 PROX2prospero homeobox 2 (ENSG00000119608), score: 0.55 PSMC2proteasome (prosome, macropain) 26S subunit, ATPase, 2 (ENSG00000161057), score: 0.57 PUS3pseudouridylate synthase 3 (ENSG00000110060), score: 0.57 RAB3GAP1RAB3 GTPase activating protein subunit 1 (catalytic) (ENSG00000115839), score: -0.54 RABEPKRab9 effector protein with kelch motifs (ENSG00000136933), score: 0.55 RAG2recombination activating gene 2 (ENSG00000175097), score: 0.83 RBM27RNA binding motif protein 27 (ENSG00000091009), score: 0.51 RBM46RNA binding motif protein 46 (ENSG00000151962), score: 0.62 RBM7RNA binding motif protein 7 (ENSG00000076053), score: 0.65 RFX3regulatory factor X, 3 (influences HLA class II expression) (ENSG00000080298), score: 0.54 RFX6regulatory factor X, 6 (ENSG00000185002), score: 0.93 RHOrhodopsin (ENSG00000163914), score: 0.54 RIBC2RIB43A domain with coiled-coils 2 (ENSG00000128408), score: 0.55 RNF17ring finger protein 17 (ENSG00000132972), score: 0.67 RNF32ring finger protein 32 (ENSG00000105982), score: 0.54 RNF38ring finger protein 38 (ENSG00000137075), score: 0.64 ROPN1Lropporin 1-like (ENSG00000145491), score: 0.54 RRM1ribonucleotide reductase M1 (ENSG00000167325), score: 0.53 RSBN1round spermatid basic protein 1 (ENSG00000081019), score: 0.6 RSPH1radial spoke head 1 homolog (Chlamydomonas) (ENSG00000160188), score: 0.69 RSPH4Aradial spoke head 4 homolog A (Chlamydomonas) (ENSG00000111834), score: 0.51 SAP30LSAP30-like (ENSG00000164576), score: -0.49 SASS6spindle assembly 6 homolog (C. elegans) (ENSG00000156876), score: 0.62 SAV1salvador homolog 1 (Drosophila) (ENSG00000151748), score: 0.54 SCARB2scavenger receptor class B, member 2 (ENSG00000138760), score: -0.53 SEC23IPSEC23 interacting protein (ENSG00000107651), score: 0.52 SGK1serum/glucocorticoid regulated kinase 1 (ENSG00000118515), score: -0.58 SH2D3CSH2 domain containing 3C (ENSG00000095370), score: -0.51 SH3GLB1SH3-domain GRB2-like endophilin B1 (ENSG00000097033), score: 0.53 SHCBP1SHC SH2-domain binding protein 1 (ENSG00000171241), score: 0.63 SLC25A29solute carrier family 25, member 29 (ENSG00000197119), score: -0.59 SMC1Bstructural maintenance of chromosomes 1B (ENSG00000077935), score: 0.61 SMC5structural maintenance of chromosomes 5 (ENSG00000198887), score: 0.58 SPACA1sperm acrosome associated 1 (ENSG00000118434), score: 0.63 SPATA18spermatogenesis associated 18 homolog (rat) (ENSG00000163071), score: 0.61 SPATA4spermatogenesis associated 4 (ENSG00000150628), score: 0.57 SPC25SPC25, NDC80 kinetochore complex component, homolog (S. cerevisiae) (ENSG00000152253), score: 0.58 SPO11SPO11 meiotic protein covalently bound to DSB homolog (S. cerevisiae) (ENSG00000054796), score: 0.91 SPTBN1spectrin, beta, non-erythrocytic 1 (ENSG00000115306), score: -0.55 SRIsorcin (ENSG00000075142), score: -0.59 SSH2slingshot homolog 2 (Drosophila) (ENSG00000141298), score: 0.5 STRA8stimulated by retinoic acid gene 8 homolog (mouse) (ENSG00000146857), score: 0.75 SYCP1synaptonemal complex protein 1 (ENSG00000198765), score: 0.6 TALDO1transaldolase 1 (ENSG00000177156), score: -0.58 TCTEX1D1Tctex1 domain containing 1 (ENSG00000152760), score: 0.58 TDP1tyrosyl-DNA phosphodiesterase 1 (ENSG00000042088), score: 0.69 TDRD1tudor domain containing 1 (ENSG00000095627), score: 0.6 TDRD5tudor domain containing 5 (ENSG00000162782), score: 0.56 TDRD6tudor domain containing 6 (ENSG00000180113), score: 0.63 TEKT3tektin 3 (ENSG00000125409), score: 0.68 TEKT5tektin 5 (ENSG00000153060), score: 0.54 TEX10testis expressed 10 (ENSG00000136891), score: 0.59 THAP1THAP domain containing, apoptosis associated protein 1 (ENSG00000131931), score: 0.54 TLX1T-cell leukemia homeobox 1 (ENSG00000107807), score: 0.98 TMC1transmembrane channel-like 1 (ENSG00000165091), score: 0.54 TMEM156transmembrane protein 156 (ENSG00000121895), score: 0.73 TMEM63Btransmembrane protein 63B (ENSG00000137216), score: -0.64 TMF1TATA element modulatory factor 1 (ENSG00000144747), score: 0.56 TOP2Atopoisomerase (DNA) II alpha 170kDa (ENSG00000131747), score: 0.61 TP53TG5TP53 target 5 (ENSG00000124251), score: 0.61 TP63tumor protein p63 (ENSG00000073282), score: 0.63 TRIB2tribbles homolog 2 (Drosophila) (ENSG00000071575), score: -0.54 TRIM36tripartite motif-containing 36 (ENSG00000152503), score: 0.54 TRIM71tripartite motif-containing 71 (ENSG00000206557), score: 0.52 TRIM8tripartite motif-containing 8 (ENSG00000171206), score: -0.56 TSC22D3TSC22 domain family, member 3 (ENSG00000157514), score: -0.52 TSGA14testis specific, 14 (ENSG00000106477), score: 0.68 TSPAN9tetraspanin 9 (ENSG00000011105), score: -0.51 TTC21Btetratricopeptide repeat domain 21B (ENSG00000123607), score: 0.59 TTC25tetratricopeptide repeat domain 25 (ENSG00000204815), score: 0.68 TTC29tetratricopeptide repeat domain 29 (ENSG00000137473), score: 0.51 TTKTTK protein kinase (ENSG00000112742), score: 0.52 TXNDC9thioredoxin domain containing 9 (ENSG00000115514), score: 0.52 UHRF1ubiquitin-like with PHD and ring finger domains 1 (ENSG00000034063), score: 0.7 URB2URB2 ribosome biogenesis 2 homolog (S. cerevisiae) (ENSG00000135763), score: 0.62 USP44ubiquitin specific peptidase 44 (ENSG00000136014), score: 0.5 USP47ubiquitin specific peptidase 47 (ENSG00000170242), score: 0.52 USP7ubiquitin specific peptidase 7 (herpes virus-associated) (ENSG00000187555), score: 0.51 VASH2vasohibin 2 (ENSG00000143494), score: 0.77 WDFY1WD repeat and FYVE domain containing 1 (ENSG00000085449), score: 0.51 WDR16WD repeat domain 16 (ENSG00000166596), score: 0.61 WDR64WD repeat domain 64 (ENSG00000162843), score: 0.65 WDR78WD repeat domain 78 (ENSG00000152763), score: 0.61 WIPF1WAS/WASL interacting protein family, member 1 (ENSG00000115935), score: -0.51 YOD1YOD1 OTU deubiquinating enzyme 1 homolog (S. cerevisiae) (ENSG00000180667), score: 0.58 ZCCHC2zinc finger, CCHC domain containing 2 (ENSG00000141664), score: 0.64 ZDHHC18zinc finger, DHHC-type containing 18 (ENSG00000204160), score: -0.51 ZFAND3zinc finger, AN1-type domain 3 (ENSG00000156639), score: 0.55 ZMYND11zinc finger, MYND domain containing 11 (ENSG00000015171), score: -0.54 ZNRF4zinc and ring finger 4 (ENSG00000105428), score: 0.69 ZPBPzona pellucida binding protein (ENSG00000042813), score: 0.62 ZPBP2zona pellucida binding protein 2 (ENSG00000186075), score: 0.57
| Id | species | tissue | sex | individual |
|---|---|---|---|---|
| gga_ts_m1_ca1 | gga | ts | m | 1 |
| gga_ts_m2_ca1 | gga | ts | m | 2 |
| mmu_ts_m1_ca1 | mmu | ts | m | 1 |
| mmu_ts_m2_ca1 | mmu | ts | m | 2 |