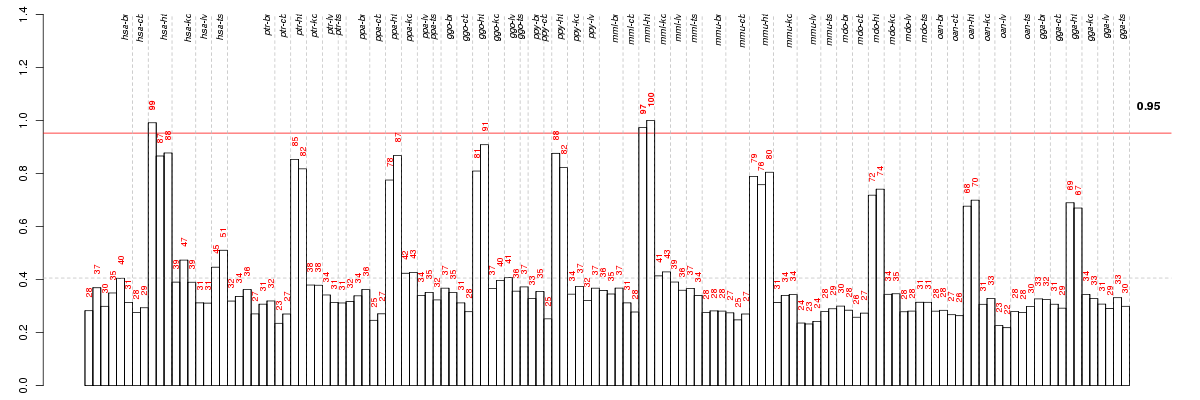

Under-expression is coded with green,
over-expression with red color.

metabolic process
The chemical reactions and pathways, including anabolism and catabolism, by which living organisms transform chemical substances. Metabolic processes typically transform small molecules, but also include macromolecular processes such as DNA repair and replication, and protein synthesis and degradation.
regulation of the force of heart contraction
Any process that modulates the extent of heart contraction, changing the force with which blood is propelled.
heart morphogenesis
The developmental process by which the heart is generated and organized. The heart is a hollow, muscular organ, which, by contracting rhythmically, keeps up the circulation of the blood.
system process
A multicellular organismal process carried out by any of the organs or tissues in an organ system. An organ system is a regularly interacting or interdependent group of organs or tissues that work together to carry out a biological objective.
muscle system process
A organ system process carried out at the level of a muscle. Muscle tissue is composed of contractile cells or fibers.
circulatory system process
A organ system process carried out by any of the organs or tissues of the circulatory system. The circulatory system is an organ system that moves extracellular fluids to and from tissue within a multicellular organism.
heart process
A circulatory system process carried out by the heart. The heart is a hollow, muscular organ, which, by contracting rhythmically, keeps up the circulation of the blood. The heart is a hollow, muscular organ, which, by contracting rhythmically, keeps up the circulation of the blood.
cardiac chamber development
The progression of a cardiac chamber over time, from its formation to the mature structure. A cardiac chamber is an enclosed cavity within the heart.
cardiac chamber morphogenesis
The process by which a cardiac chamber is generated and organized. A cardiac chamber is an enclosed cavity within the heart.
cardiac ventricle morphogenesis
The process by which the cardiac ventricle is generated and organized. A cardiac ventricle receives blood from a cardiac atrium and pumps it out of the heart.
ventricular cardiac muscle tissue development
The process whose specific outcome is the progression of ventricular cardiac muscle over time, from its formation to the mature structure.
cardiac ventricle development
The process whose specific outcome is the progression of a cardiac ventricle over time, from its formation to the mature structure. A cardiac ventricle receives blood from a cardiac atrium and pumps it out of the heart.
muscle contraction
A process whereby force is generated within muscle tissue, resulting in a change in muscle geometry. Force generation involves a chemo-mechanical energy conversion step that is carried out by the actin/myosin complex activity, which generates force through ATP hydrolysis.
actin cytoskeleton organization
A process that is carried out at the cellular level which results in the assembly, arrangement of constituent parts, or disassembly of cytoskeletal structures comprising actin filaments and their associated proteins.
cytoskeleton organization
A process that is carried out at the cellular level which results in the assembly, arrangement of constituent parts, or disassembly of cytoskeletal structures.
nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
Any cellular metabolic process involving nucleobases, nucleosides, nucleotides and nucleic acids.
regulation of nucleotide metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways involving nucleotides.
purine nucleotide metabolic process
The chemical reactions and pathways involving a purine nucleotide, a compound consisting of nucleoside (a purine base linked to a deoxyribose or ribose sugar) esterified with a phosphate moiety at either the 3' or 5'-hydroxyl group of its glycose moiety.
purine nucleotide catabolic process
The chemical reactions and pathways resulting in the breakdown of a purine nucleotide, a compound consisting of nucleoside (a purine base linked to a deoxyribose or ribose sugar) esterified with a phosphate moiety at either the 3' or 5'-hydroxyl group of its glycose moiety.
protein modification process
The covalent alteration of one or more amino acids occurring in proteins, peptides and nascent polypeptides (co-translational, post-translational modifications). Includes the modification of charged tRNAs that are destined to occur in a protein (pre-translation modification).
nucleoside phosphate metabolic process
The chemical reactions and pathways involving any phosphorylated nucleoside.
nitrogen compound metabolic process
The chemical reactions and pathways involving various organic and inorganic nitrogenous compounds; includes nitrogen fixation, nitrification, denitrification, assimilatory/dissimilatory nitrate reduction and the interconversion of nitrogenous organic matter and ammonium.
transport
The directed movement of substances (such as macromolecules, small molecules, ions) into, out of, within or between cells, or within a multicellular organism by means of some external agent such as a transporter or pore.
striated muscle contraction
A process whereby force is generated within striated muscle tissue, resulting in the shortening of the muscle. Force generation involves a chemo-mechanical energy conversion step that is carried out by the actin/myosin complex activity, which generates force through ATP hydrolysis. Striated muscle is a type of muscle in which the repeating units (sarcomeres) of the contractile myofibrils are arranged in registry throughout the cell, resulting in transverse or oblique striations observable at the level of the light microscope.
organelle organization
A process that is carried out at the cellular level which results in the assembly, arrangement of constituent parts, or disassembly of an organelle within a cell. An organelle is an organized structure of distinctive morphology and function. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton. Excludes the plasma membrane.
multicellular organismal development
The biological process whose specific outcome is the progression of a multicellular organism over time from an initial condition (e.g. a zygote or a young adult) to a later condition (e.g. a multicellular animal or an aged adult).
anatomical structure morphogenesis
The process by which anatomical structures are generated and organized. Morphogenesis pertains to the creation of form.
organ morphogenesis
Morphogenesis of an organ. An organ is defined as a tissue or set of tissues that work together to perform a specific function or functions. Morphogenesis is the process by which anatomical structures are generated and organized. Organs are commonly observed as visibly distinct structures, but may also exist as loosely associated clusters of cells that work together to perform a specific function or functions.
tissue development
The process whose specific outcome is the progression of a tissue over time, from its formation to the mature structure.
heart development
The process whose specific outcome is the progression of the heart over time, from its formation to the mature structure. The heart is a hollow, muscular organ, which, by contracting rhythmically, keeps up the circulation of the blood.
muscle organ development
The process whose specific outcome is the progression of the muscle over time, from its formation to the mature structure. The muscle is an organ consisting of a tissue made up of various elongated cells that are specialized to contract and thus to produce movement and mechanical work.
blood circulation
The flow of blood through the body of an animal, enabling the transport of nutrients to the tissues and the removal of waste products.
regulation of heart contraction
Any process that modulates the frequency, rate or extent of heart contraction. Heart contraction is the process by which the heart decreases in volume in a characteristic way to propel blood through the body.
biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
actin filament-based movement
Movement of organelles or other particles along actin filaments, or sliding of actin filaments past each other, mediated by motor proteins.
catabolic process
The chemical reactions and pathways resulting in the breakdown of substances, including the breakdown of carbon compounds with the liberation of energy for use by the cell or organism.
nucleotide metabolic process
The chemical reactions and pathways involving a nucleotide, a nucleoside that is esterified with (ortho)phosphate or an oligophosphate at any hydroxyl group on the glycose moiety; may be mono-, di- or triphosphate; this definition includes cyclic nucleotides (nucleoside cyclic phosphates).
nucleotide catabolic process
The chemical reactions and pathways resulting in the breakdown of nucleotides, any nucleoside that is esterified with (ortho)phosphate or an oligophosphate at any hydroxyl group on the glycose moiety; may be mono-, di- or triphosphate; this definition includes cyclic-nucleotides (nucleoside cyclic phosphates).
regulation of catabolic process
Any process that modulates the frequency, rate, or extent of the chemical reactions and pathways resulting in the breakdown of substances.
cellular process
Any process that is carried out at the cellular level, but not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
cellular component assembly
The aggregation, arrangement and bonding together of a cellular component.
anatomical structure formation involved in morphogenesis
The developmental process pertaining to the initial formation of an anatomical structure from unspecified parts. This process begins with the specific processes that contribute to the appearance of the discrete structure and ends when the structural rudiment is recognizable. An anatomical structure is any biological entity that occupies space and is distinguished from its surroundings. Anatomical structures can be macroscopic such as a carpel, or microscopic such as an acrosome.
cellular component assembly involved in morphogenesis
The cellular component assembly that is part of the initial shaping of the component during its developmental progression.
striated muscle tissue development
The process whose specific outcome is the progression of a striated muscle over time, from its formation to the mature structure. Striated muscle contain fibers that are divided by transverse bands into striations, and cardiac and skeletal muscle are types of striated muscle. Skeletal muscle myoblasts fuse to form myotubes and eventually multinucleated muscle fibers. The fusion of cardiac cells is very rare and can only form binucleate cells.
cellular component organization
A process that is carried out at the cellular level which results in the assembly, arrangement of constituent parts, or disassembly of a cellular component.
peptide cross-linking
The formation of a covalent cross-link between or within protein chains.
regulation of nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
Any cellular process that modulates the frequency, rate or extent of the chemical reactions and pathways involving nucleobases, nucleosides, nucleotides and nucleic acids.
regulation of metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways within a cell or an organism.
protein metabolic process
The chemical reactions and pathways involving a specific protein, rather than of proteins in general. Includes protein modification.
actin filament-based process
Any cellular process that depends upon or alters the actin cytoskeleton, that part of the cytoskeleton comprising actin filaments and their associated proteins.
muscle filament sliding
The sliding of actin thin filaments and myosin thick filaments past each other in muscle contraction. This involves a process of interaction of myosin located on a thick filament with actin located on a thin filament. During this process ATP is split and forces are generated.
cell differentiation
The process whereby relatively unspecialized cells, e.g. embryonic or regenerative cells, acquire specialized structural and/or functional features that characterize the cells, tissues, or organs of the mature organism or some other relatively stable phase of the organism's life history. Differentiation includes the processes involved in commitment of a cell to a specific fate and its subsequent development to the mature state.
extracellular matrix organization
A process that is carried out at the cellular level which results in the assembly, arrangement of constituent parts, or disassembly of an extracellular matrix.
myofibril assembly
Formation of myofibrils, the repeating units of striated muscle.
cytoskeleton-dependent intracellular transport
The directed movement of substances along cytoskeletal elements such as microfilaments or microtubules within a cell.
regulation of nucleotide catabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways resulting in the breakdown of nucleotides.
actomyosin structure organization
A process that is carried out at the cellular level which results in the assembly, arrangement of constituent parts, or disassembly of cytoskeletal structures containing both actin and myosin or paramyosin. The myosin may be organized into filaments.
regulation of cellular metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways by which individual cells transform chemical substances.
regulation of cellular catabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways resulting in the breakdown of substances, carried out by individual cells.
multicellular organismal process
Any biological process, occurring at the level of a multicellular organism, pertinent to its function.
developmental process
A biological process whose specific outcome is the progression of an integrated living unit: an anatomical structure (which may be a subcellular structure, cell, tissue, or organ), or organism over time from an initial condition to a later condition.
positive regulation of ATPase activity
Any process that activates or increases the rate of ATP hydrolysis by an ATPase.
cellular component morphogenesis
The process by which cellular structures, including whole cells or cell parts, are generated and organized. Morphogenesis pertains to the creation of form.
regulation of purine nucleotide catabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways resulting in the breakdown of purine nucleotides.
actin-myosin filament sliding
The sliding movement of actin thin filaments and myosin thick filaments past each other.
cellular nitrogen compound metabolic process
The chemical reactions and pathways involving various organic and inorganic nitrogenous compounds, as carried out by individual cells.
nucleobase, nucleoside, nucleotide and nucleic acid catabolic process
The chemical reactions and pathways resulting in the breakdown of nucleobases, nucleosides, nucleotides and nucleic acids.
nucleobase, nucleoside and nucleotide catabolic process
The chemical reactions and pathways resulting in the breakdown of nucleobases, nucleosides and nucleotides.
cardiac cell differentiation
The process whereby a relatively unspecialized cell acquires the specialized structural and/or functional features of a cell that will form part of the cardiac organ of an individual.
muscle cell differentiation
The process whereby a relatively unspecialized cell acquires specialized features of a muscle cell.
extracellular structure organization
A process that is carried out at the cellular level which results in the assembly, arrangement of constituent parts, or disassembly of structures in the space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane, and also covers the host cell environment outside an intracellular parasite.
positive regulation of catalytic activity
Any process that activates or increases the activity of an enzyme.
macromolecule metabolic process
The chemical reactions and pathways involving macromolecules, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass.
macromolecule modification
The covalent alteration of one or more monomeric units in a polypeptide, polynucleotide, polysaccharide, or other biological macromolecule, resulting in a change in its properties.
regulation of ATPase activity
Any process that modulates the rate of ATP hydrolysis by an ATPase.
post-translational protein modification
The covalent alteration of one or more amino acids occurring in a protein after the protein has been completely translated and released from the ribosome.
regulation of system process
Any process that modulates the frequency, rate or extent of a system process, a multicellular organismal process carried out by any of the organs or tissues in an organ system.
cellular component biogenesis
The process by which a cellular component is synthesized, aggregates, and bonds together. Includes biosynthesis of constituent macromolecules, and those macromolecular modifications that are involved in synthesis or assembly of the cellular component.
positive regulation of molecular function
Any process that activates or increases the rate or extent of a molecular function, an elemental biological activity occurring at the molecular level, such as catalysis or binding.
cellular metabolic process
The chemical reactions and pathways by which individual cells transform chemical substances.
primary metabolic process
The chemical reactions and pathways involving those compounds which are formed as a part of the normal anabolic and catabolic processes. These processes take place in most, if not all, cells of the organism.
cellular catabolic process
The chemical reactions and pathways resulting in the breakdown of substances, carried out by individual cells.
cellular macromolecule metabolic process
The chemical reactions and pathways involving macromolecules, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass, as carried out by individual cells.
cellular protein metabolic process
The chemical reactions and pathways involving a specific protein, rather than of proteins in general, occurring at the level of an individual cell. Includes protein modification.
cellular nitrogen compound catabolic process
The chemical reactions and pathways resulting in the breakdown of organic and inorganic nitrogenous compounds.
small molecule metabolic process
The chemical reactions and pathways involving small molecules, any monomeric molecule of small relative molecular mass.
small molecule catabolic process
The chemical reactions and pathways resulting in the breakdown of small molecules, any monomeric molecule of small relative molecular mass.
heterocycle metabolic process
The chemical reactions and pathways involving heterocyclic compounds, those with a cyclic molecular structure and at least two different atoms in the ring (or rings).
heterocycle catabolic process
The chemical reactions and pathways resulting in the breakdown of heterocyclic compounds, those with a cyclic molecular structure and at least two different atoms in the ring (or rings).
intracellular transport
The directed movement of substances within a cell.
cell development
The process whose specific outcome is the progression of the cell over time, from its formation to the mature structure. Cell development does not include the steps involved in committing a cell to a specific fate.
organ development
Development of a tissue or tissues that work together to perform a specific function or functions. Development pertains to the process whose specific outcome is the progression of a structure over time, from its formation to the mature structure. Organs are commonly observed as visibly distinct structures, but may also exist as loosely associated clusters of cells that work together to perform a specific function or functions.
tissue morphogenesis
The process by which the anatomical structures of a tissue are generated and organized. Morphogenesis pertains to the creation of form.
system development
The process whose specific outcome is the progression of an organismal system over time, from its formation to the mature structure. A system is a regularly interacting or interdependent group of organs or tissues that work together to carry out a given biological process.
cardiac muscle tissue development
The process whose specific outcome is the progression of cardiac muscle over time, from its formation to the mature structure.
anatomical structure development
The biological process whose specific outcome is the progression of an anatomical structure from an initial condition to its mature state. This process begins with the formation of the structure and ends with the mature structure, whatever form that may be including its natural destruction. An anatomical structure is any biological entity that occupies space and is distinguished from its surroundings. Anatomical structures can be macroscopic such as a carpel, or microscopic such as an acrosome.
cellular developmental process
A biological process whose specific outcome is the progression of a cell over time from an initial condition to a later condition.
regulation of biological process
Any process that modulates the frequency, rate or extent of a biological process. Biological processes are regulated by many means; examples include the control of gene expression, protein modification or interaction with a protein or substrate molecule.
regulation of catalytic activity
Any process that modulates the activity of an enzyme.
regulation of cellular process
Any process that modulates the frequency, rate or extent of a cellular process, any of those that are carried out at the cellular level, but are not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
striated muscle cell differentiation
The process whereby a relatively unspecialized cell acquires specialized features of a striated muscle cell; striated muscle fibers are divided by transverse bands into striations, and cardiac and voluntary muscle are types of striated muscle.
regulation of nitrogen compound metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways involving nitrogen or nitrogenous compounds.
localization
Any process by which a cell, a substance, or a cellular entity, such as a protein complex or organelle, is transported to, and/or maintained in a specific location.
establishment of localization
The directed movement of a cell, substance or cellular entity, such as a protein complex or organelle, to a specific location.
regulation of multicellular organismal process
Any process that modulates the frequency, rate or extent of a multicellular organismal process, the processes pertinent to the function of a multicellular organism above the cellular level; includes the integrated processes of tissues and organs.
regulation of hydrolase activity
Any process that modulates the frequency, rate or extent of hydrolase activity, the catalysis of the hydrolysis of various bonds, e.g. C-O, C-N, C-C, phosphoric anhydride bonds, etc. Hydrolase is the systematic name for any enzyme of EC class 3.
positive regulation of hydrolase activity
Any process that activates or increases the frequency, rate or extent of hydrolase activity, the catalysis of the hydrolysis of various bonds.
cellular localization
Any process by which a substance or cellular entity, such as a protein complex or organelle, is transported to, and/or maintained in, a specific location within or in the membrane of a cell.
establishment of localization in cell
The directed movement of a substance or cellular entity, such as a protein complex or organelle, to a specific location within, or in the membrane of, a cell.
muscle cell development
The process whose specific outcome is the progression of a muscle cell over time, from its formation to the mature structure. Muscle cell development does not include the steps involved in committing an unspecified cell to the muscle cell fate.
striated muscle cell development
The process whose specific outcome is the progression of a striated muscle cell over time, from its formation to the mature structure. Striated muscle cells contain fibers that are divided by transverse bands into striations, and cardiac and skeletal muscle are types of striated muscle.
cardiac muscle tissue morphogenesis
The process by which the anatomical structures of cardiac muscle tissue are generated and organized. Morphogenesis pertains to the creation of form.
ventricular cardiac muscle tissue morphogenesis
The process by which the anatomical structures of cardiac ventricle muscle is generated and organized. Morphogenesis pertains to the creation of form.
nucleobase, nucleoside and nucleotide metabolic process
The cellular chemical reactions and pathways involving nucleobases, nucleosides and nucleotides.
heart contraction
The multicellular organismal process by which the heart decreases in volume in a characteristic way to propel blood through the body.
cardiac muscle contraction
Muscle contraction of cardiac muscle tissue.
muscle tissue morphogenesis
The process by which the anatomical structures of muscle tissue are generated and organized. Muscle tissue consists of a set of cells that are part of an organ and carry out a contractive function. Morphogenesis pertains to the creation of form.
muscle tissue development
The progression of muscle tissue over time, from its initial formation to its mature state. Muscle tissue is a contractile tissue made up of actin and myosin fibers.
muscle structure development
The progression of a muscle structure over time, from its formation to its mature state. Muscle structures are contractile cells, tissues or organs that are found in multicellular organisms.
biological regulation
Any process that modulates the frequency, rate or extent of any biological process, quality or function.
regulation of biological quality
Any process that modulates the frequency, rate or extent of a biological quality. A biological quality is a measurable attribute of an organism or part of an organism, such as size, mass, shape, color, etc.
regulation of molecular function
Any process that modulates the frequency, rate or extent of a molecular function, an elemental biological activity occurring at the molecular level, such as catalysis or binding.
actin-mediated cell contraction
The actin filament-based process by which cytoplasmic actin filaments slide past one another resulting in contraction of all or part of the cell body.
regulation of primary metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways within a cell or an organism involving those compounds formed as a part of the normal anabolic and catabolic processes. These processes take place in most, if not all, cells of the organism.
all
NA
cellular metabolic process
The chemical reactions and pathways by which individual cells transform chemical substances.
organelle organization
A process that is carried out at the cellular level which results in the assembly, arrangement of constituent parts, or disassembly of an organelle within a cell. An organelle is an organized structure of distinctive morphology and function. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton. Excludes the plasma membrane.
extracellular structure organization
A process that is carried out at the cellular level which results in the assembly, arrangement of constituent parts, or disassembly of structures in the space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane, and also covers the host cell environment outside an intracellular parasite.
multicellular organismal development
The biological process whose specific outcome is the progression of a multicellular organism over time from an initial condition (e.g. a zygote or a young adult) to a later condition (e.g. a multicellular animal or an aged adult).
cellular developmental process
A biological process whose specific outcome is the progression of a cell over time from an initial condition to a later condition.
cellular component assembly
The aggregation, arrangement and bonding together of a cellular component.
regulation of metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways within a cell or an organism.
regulation of cellular process
Any process that modulates the frequency, rate or extent of a cellular process, any of those that are carried out at the cellular level, but are not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
regulation of multicellular organismal process
Any process that modulates the frequency, rate or extent of a multicellular organismal process, the processes pertinent to the function of a multicellular organism above the cellular level; includes the integrated processes of tissues and organs.
establishment of localization
The directed movement of a cell, substance or cellular entity, such as a protein complex or organelle, to a specific location.
cellular localization
Any process by which a substance or cellular entity, such as a protein complex or organelle, is transported to, and/or maintained in, a specific location within or in the membrane of a cell.
regulation of biological process
Any process that modulates the frequency, rate or extent of a biological process. Biological processes are regulated by many means; examples include the control of gene expression, protein modification or interaction with a protein or substrate molecule.
regulation of catabolic process
Any process that modulates the frequency, rate, or extent of the chemical reactions and pathways resulting in the breakdown of substances.
regulation of nitrogen compound metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways involving nitrogen or nitrogenous compounds.
regulation of cellular metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways by which individual cells transform chemical substances.
cellular nitrogen compound metabolic process
The chemical reactions and pathways involving various organic and inorganic nitrogenous compounds, as carried out by individual cells.
cellular catabolic process
The chemical reactions and pathways resulting in the breakdown of substances, carried out by individual cells.
cellular macromolecule metabolic process
The chemical reactions and pathways involving macromolecules, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass, as carried out by individual cells.
protein metabolic process
The chemical reactions and pathways involving a specific protein, rather than of proteins in general. Includes protein modification.
regulation of primary metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways within a cell or an organism involving those compounds formed as a part of the normal anabolic and catabolic processes. These processes take place in most, if not all, cells of the organism.
small molecule catabolic process
The chemical reactions and pathways resulting in the breakdown of small molecules, any monomeric molecule of small relative molecular mass.
cellular component morphogenesis
The process by which cellular structures, including whole cells or cell parts, are generated and organized. Morphogenesis pertains to the creation of form.
regulation of cellular metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways by which individual cells transform chemical substances.
establishment of localization in cell
The directed movement of a substance or cellular entity, such as a protein complex or organelle, to a specific location within, or in the membrane of, a cell.
cellular component assembly involved in morphogenesis
The cellular component assembly that is part of the initial shaping of the component during its developmental progression.
regulation of system process
Any process that modulates the frequency, rate or extent of a system process, a multicellular organismal process carried out by any of the organs or tissues in an organ system.
anatomical structure formation involved in morphogenesis
The developmental process pertaining to the initial formation of an anatomical structure from unspecified parts. This process begins with the specific processes that contribute to the appearance of the discrete structure and ends when the structural rudiment is recognizable. An anatomical structure is any biological entity that occupies space and is distinguished from its surroundings. Anatomical structures can be macroscopic such as a carpel, or microscopic such as an acrosome.
cellular component morphogenesis
The process by which cellular structures, including whole cells or cell parts, are generated and organized. Morphogenesis pertains to the creation of form.
cellular component assembly involved in morphogenesis
The cellular component assembly that is part of the initial shaping of the component during its developmental progression.
anatomical structure morphogenesis
The process by which anatomical structures are generated and organized. Morphogenesis pertains to the creation of form.
cell development
The process whose specific outcome is the progression of the cell over time, from its formation to the mature structure. Cell development does not include the steps involved in committing a cell to a specific fate.
system development
The process whose specific outcome is the progression of an organismal system over time, from its formation to the mature structure. A system is a regularly interacting or interdependent group of organs or tissues that work together to carry out a given biological process.
intracellular transport
The directed movement of substances within a cell.
regulation of catalytic activity
Any process that modulates the activity of an enzyme.
nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
Any cellular metabolic process involving nucleobases, nucleosides, nucleotides and nucleic acids.
regulation of cellular catabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways resulting in the breakdown of substances, carried out by individual cells.
cellular nitrogen compound catabolic process
The chemical reactions and pathways resulting in the breakdown of organic and inorganic nitrogenous compounds.
regulation of nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
Any cellular process that modulates the frequency, rate or extent of the chemical reactions and pathways involving nucleobases, nucleosides, nucleotides and nucleic acids.
regulation of cellular catabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways resulting in the breakdown of substances, carried out by individual cells.
regulation of nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
Any cellular process that modulates the frequency, rate or extent of the chemical reactions and pathways involving nucleobases, nucleosides, nucleotides and nucleic acids.
cellular protein metabolic process
The chemical reactions and pathways involving a specific protein, rather than of proteins in general, occurring at the level of an individual cell. Includes protein modification.
heterocycle catabolic process
The chemical reactions and pathways resulting in the breakdown of heterocyclic compounds, those with a cyclic molecular structure and at least two different atoms in the ring (or rings).
regulation of nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
Any cellular process that modulates the frequency, rate or extent of the chemical reactions and pathways involving nucleobases, nucleosides, nucleotides and nucleic acids.
nucleobase, nucleoside and nucleotide metabolic process
The cellular chemical reactions and pathways involving nucleobases, nucleosides and nucleotides.
nucleobase, nucleoside and nucleotide catabolic process
The chemical reactions and pathways resulting in the breakdown of nucleobases, nucleosides and nucleotides.
actin cytoskeleton organization
A process that is carried out at the cellular level which results in the assembly, arrangement of constituent parts, or disassembly of cytoskeletal structures comprising actin filaments and their associated proteins.
actin-myosin filament sliding
The sliding movement of actin thin filaments and myosin thick filaments past each other.
cell development
The process whose specific outcome is the progression of the cell over time, from its formation to the mature structure. Cell development does not include the steps involved in committing a cell to a specific fate.
organ development
Development of a tissue or tissues that work together to perform a specific function or functions. Development pertains to the process whose specific outcome is the progression of a structure over time, from its formation to the mature structure. Organs are commonly observed as visibly distinct structures, but may also exist as loosely associated clusters of cells that work together to perform a specific function or functions.
tissue morphogenesis
The process by which the anatomical structures of a tissue are generated and organized. Morphogenesis pertains to the creation of form.
cardiac chamber morphogenesis
The process by which a cardiac chamber is generated and organized. A cardiac chamber is an enclosed cavity within the heart.
tissue morphogenesis
The process by which the anatomical structures of a tissue are generated and organized. Morphogenesis pertains to the creation of form.
organ morphogenesis
Morphogenesis of an organ. An organ is defined as a tissue or set of tissues that work together to perform a specific function or functions. Morphogenesis is the process by which anatomical structures are generated and organized. Organs are commonly observed as visibly distinct structures, but may also exist as loosely associated clusters of cells that work together to perform a specific function or functions.
tissue development
The process whose specific outcome is the progression of a tissue over time, from its formation to the mature structure.
muscle organ development
The process whose specific outcome is the progression of the muscle over time, from its formation to the mature structure. The muscle is an organ consisting of a tissue made up of various elongated cells that are specialized to contract and thus to produce movement and mechanical work.
muscle cell differentiation
The process whereby a relatively unspecialized cell acquires specialized features of a muscle cell.
positive regulation of catalytic activity
Any process that activates or increases the activity of an enzyme.
nucleobase, nucleoside, nucleotide and nucleic acid catabolic process
The chemical reactions and pathways resulting in the breakdown of nucleobases, nucleosides, nucleotides and nucleic acids.
positive regulation of hydrolase activity
Any process that activates or increases the frequency, rate or extent of hydrolase activity, the catalysis of the hydrolysis of various bonds.
protein modification process
The covalent alteration of one or more amino acids occurring in proteins, peptides and nascent polypeptides (co-translational, post-translational modifications). Includes the modification of charged tRNAs that are destined to occur in a protein (pre-translation modification).
purine nucleotide catabolic process
The chemical reactions and pathways resulting in the breakdown of a purine nucleotide, a compound consisting of nucleoside (a purine base linked to a deoxyribose or ribose sugar) esterified with a phosphate moiety at either the 3' or 5'-hydroxyl group of its glycose moiety.
nucleobase, nucleoside and nucleotide catabolic process
The chemical reactions and pathways resulting in the breakdown of nucleobases, nucleosides and nucleotides.
myofibril assembly
Formation of myofibrils, the repeating units of striated muscle.
muscle cell development
The process whose specific outcome is the progression of a muscle cell over time, from its formation to the mature structure. Muscle cell development does not include the steps involved in committing an unspecified cell to the muscle cell fate.
muscle filament sliding
The sliding of actin thin filaments and myosin thick filaments past each other in muscle contraction. This involves a process of interaction of myosin located on a thick filament with actin located on a thin filament. During this process ATP is split and forces are generated.
heart contraction
The multicellular organismal process by which the heart decreases in volume in a characteristic way to propel blood through the body.
regulation of the force of heart contraction
Any process that modulates the extent of heart contraction, changing the force with which blood is propelled.
cardiac chamber morphogenesis
The process by which a cardiac chamber is generated and organized. A cardiac chamber is an enclosed cavity within the heart.
cardiac muscle tissue morphogenesis
The process by which the anatomical structures of cardiac muscle tissue are generated and organized. Morphogenesis pertains to the creation of form.
cardiac ventricle morphogenesis
The process by which the cardiac ventricle is generated and organized. A cardiac ventricle receives blood from a cardiac atrium and pumps it out of the heart.
heart morphogenesis
The developmental process by which the heart is generated and organized. The heart is a hollow, muscular organ, which, by contracting rhythmically, keeps up the circulation of the blood.
cardiac chamber development
The progression of a cardiac chamber over time, from its formation to the mature structure. A cardiac chamber is an enclosed cavity within the heart.
cardiac cell differentiation
The process whereby a relatively unspecialized cell acquires the specialized structural and/or functional features of a cell that will form part of the cardiac organ of an individual.
muscle tissue development
The progression of muscle tissue over time, from its initial formation to its mature state. Muscle tissue is a contractile tissue made up of actin and myosin fibers.
actin filament-based movement
Movement of organelles or other particles along actin filaments, or sliding of actin filaments past each other, mediated by motor proteins.
regulation of nucleotide catabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways resulting in the breakdown of nucleotides.
regulation of purine nucleotide catabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways resulting in the breakdown of purine nucleotides.
purine nucleotide catabolic process
The chemical reactions and pathways resulting in the breakdown of a purine nucleotide, a compound consisting of nucleoside (a purine base linked to a deoxyribose or ribose sugar) esterified with a phosphate moiety at either the 3' or 5'-hydroxyl group of its glycose moiety.
regulation of nucleotide catabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways resulting in the breakdown of nucleotides.
positive regulation of ATPase activity
Any process that activates or increases the rate of ATP hydrolysis by an ATPase.
regulation of nucleotide metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways involving nucleotides.
purine nucleotide metabolic process
The chemical reactions and pathways involving a purine nucleotide, a compound consisting of nucleoside (a purine base linked to a deoxyribose or ribose sugar) esterified with a phosphate moiety at either the 3' or 5'-hydroxyl group of its glycose moiety.
nucleotide catabolic process
The chemical reactions and pathways resulting in the breakdown of nucleotides, any nucleoside that is esterified with (ortho)phosphate or an oligophosphate at any hydroxyl group on the glycose moiety; may be mono-, di- or triphosphate; this definition includes cyclic-nucleotides (nucleoside cyclic phosphates).
striated muscle cell development
The process whose specific outcome is the progression of a striated muscle cell over time, from its formation to the mature structure. Striated muscle cells contain fibers that are divided by transverse bands into striations, and cardiac and skeletal muscle are types of striated muscle.
myofibril assembly
Formation of myofibrils, the repeating units of striated muscle.
regulation of heart contraction
Any process that modulates the frequency, rate or extent of heart contraction. Heart contraction is the process by which the heart decreases in volume in a characteristic way to propel blood through the body.
cardiac muscle contraction
Muscle contraction of cardiac muscle tissue.
ventricular cardiac muscle tissue morphogenesis
The process by which the anatomical structures of cardiac ventricle muscle is generated and organized. Morphogenesis pertains to the creation of form.
cardiac muscle tissue development
The process whose specific outcome is the progression of cardiac muscle over time, from its formation to the mature structure.
cardiac muscle tissue morphogenesis
The process by which the anatomical structures of cardiac muscle tissue are generated and organized. Morphogenesis pertains to the creation of form.
regulation of ATPase activity
Any process that modulates the rate of ATP hydrolysis by an ATPase.
ventricular cardiac muscle tissue morphogenesis
The process by which the anatomical structures of cardiac ventricle muscle is generated and organized. Morphogenesis pertains to the creation of form.

plasma membrane
The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins.
intracellular
The living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
contractile fiber
Fibers, composed of actin, myosin, and associated proteins, found in cells of smooth or striated muscle.
membrane
Double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
extracellular region
The space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers the host cell environment outside an intracellular parasite.
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
proteinaceous extracellular matrix
A layer consisting mainly of proteins (especially collagen) and glycosaminoglycans (mostly as proteoglycans) that forms a sheet underlying or overlying cells such as endothelial and epithelial cells. The proteins are secreted by cells in the vicinity. An example of this component is found in Mus musculus.
basement membrane
A thin layer of dense material found in various animal tissues interposed between the cells and the adjacent connective tissue. It consists of the basal lamina plus an associated layer of reticulin fibers.
extracellular space
That part of a multicellular organism outside the cells proper, usually taken to be outside the plasma membranes, and occupied by fluid.
cell
The basic structural and functional unit of all organisms. Includes the plasma membrane and any external encapsulating structures such as the cell wall and cell envelope.
cytoplasm
All of the contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures.
cytoskeleton
Any of the various filamentous elements that form the internal framework of cells, and typically remain after treatment of the cells with mild detergent to remove membrane constituents and soluble components of the cytoplasm. The term embraces intermediate filaments, microfilaments, microtubules, the microtrabecular lattice, and other structures characterized by a polymeric filamentous nature and long-range order within the cell. The various elements of the cytoskeleton not only serve in the maintenance of cellular shape but also have roles in other cellular functions, including cellular movement, cell division, endocytosis, and movement of organelles.
muscle myosin complex
A filament of myosin found in a muscle cell of any type.
striated muscle thin filament
Filaments formed of actin and associated proteins; attached to Z discs at either end of sarcomeres in myofibrils.
adherens junction
A cell junction at which anchoring proteins (cadherins or integrins) extend through the plasma membrane and are attached to actin filaments.
cell-substrate adherens junction
An adherens junction which connects a cell to the extracellular matrix.
actin cytoskeleton
The part of the cytoskeleton (the internal framework of a cell) composed of actin and associated proteins. Includes actin cytoskeleton-associated complexes.
basolateral plasma membrane
The region of the plasma membrane that includes the basal end and sides of the cell. Often used in reference to animal polarized epithelial membranes, where the basal membrane is the part attached to the extracellular matrix, or in plant cells, where the basal membrane is defined with respect to the zygotic axis.
myosin complex
A protein complex, formed of one or more myosin heavy chains plus associated light chains and other proteins, that functions as a molecular motor; uses the energy of ATP hydrolysis to move actin filaments or to move vesicles or other cargo on fixed actin filaments; has magnesium-ATPase activity and binds actin. Myosin classes are distinguished based on sequence features of the motor, or head, domain, but also have distinct tail regions that are believed to bind specific cargoes.
myosin II complex
A myosin complex containing two class II myosin heavy chains, two myosin essential light chains and two myosin regulatory light chains. Also known as classical myosin or conventional myosin, the myosin II class includes the major muscle myosin of vertebrate and invertebrate muscle, and is characterized by alpha-helical coiled coil tails that self assemble to form a variety of filament structures.
myofibril
The contractile element of skeletal and cardiac muscle; a long, highly organized bundle of actin, myosin, and other proteins that contracts by a sliding filament mechanism.
sarcomere
The repeating unit of a myofibril in a muscle cell, composed of an array of overlapping thick and thin filaments between two adjacent Z discs.
Z disc
Platelike region of a muscle sarcomere to which the plus ends of actin filaments are attached.
cell junction
A plasma membrane part that forms a specialized region of connection between two cells or between a cell and the extracellular matrix. At a cell junction, anchoring proteins extend through the plasma membrane to link cytoskeletal proteins in one cell to cytoskeletal proteins in neighboring cells or to proteins in the extracellular matrix.
cell-substrate junction
A cell junction that forms a connection between a cell and the extracellular matrix.
extracellular matrix
A structure lying external to one or more cells, which provides structural support for cells or tissues; may be completely external to the cell (as in animals) or be part of the cell (as in plants).
pseudopodium
A temporary protrusion or retractile process of a cell, associated with flowing movements of the protoplasm, and serving for locomotion and feeding.
A band
The dark-staining region of a sarcomere, in which myosin thick filaments are present; the center is traversed by the paler H zone, which in turn contains the M line.
I band
A region of a sarcomere that appears as a light band on each side of the Z disc, comprising a region of the sarcomere where thin (actin) filaments are not overlapped by thick (myosin) filaments; contains actin, troponin, and tropomyosin; each sarcomere includes half of an I band at each end.
macromolecular complex
A stable assembly of two or more macromolecules, i.e. proteins, nucleic acids, carbohydrates or lipids, in which the constituent parts function together.
sarcolemma
The outer membrane of a muscle cell, consisting of the plasma membrane, a covering basement membrane (about 100 nm thick and sometimes common to more than one fiber), and the associated loose network of collagen fibers.
cell projection
A prolongation or process extending from a cell, e.g. a flagellum or axon.
organelle
Organized structure of distinctive morphology and function. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton, and prokaryotic structures such as anammoxosomes and pirellulosomes. Excludes the plasma membrane.
non-membrane-bounded organelle
Organized structure of distinctive morphology and function, not bounded by a lipid bilayer membrane. Includes ribosomes, the cytoskeleton and chromosomes.
intracellular organelle
Organized structure of distinctive morphology and function, occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton. Excludes the plasma membrane.
intracellular non-membrane-bounded organelle
Organized structure of distinctive morphology and function, not bounded by a lipid bilayer membrane and occurring within the cell. Includes ribosomes, the cytoskeleton and chromosomes.
protein complex
Any macromolecular complex composed of two or more polypeptide subunits, which may or may not be identical. Protein complexes may have other associated non-protein prosthetic groups, such as nucleotides, metal ions or other small molecules.
extracellular matrix part
Any constituent part of the extracellular matrix, the structure lying external to one or more cells, which provides structural support for cells or tissues; may be completely external to the cell (as in animals) or be part of the cell (as often seen in plants).
extracellular region part
Any constituent part of the extracellular region, the space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers constituent parts of the host cell environment outside an intracellular parasite.
organelle part
Any constituent part of an organelle, an organized structure of distinctive morphology and function. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton, but excludes the plasma membrane.
intracellular part
Any constituent part of the living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
membrane part
Any constituent part of a membrane, a double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
cytoskeletal part
Any constituent part of the cytoskeleton, a cellular scaffolding or skeleton that maintains cell shape, enables some cell motion (using structures such as flagella and cilia), and plays important roles in both intra-cellular transport (e.g. the movement of vesicles and organelles) and cellular division. Includes constituent parts of intermediate filaments, microfilaments, microtubules, and the microtrabecular lattice.
cytoplasmic part
Any constituent part of the cytoplasm, all of the contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures.
intracellular organelle part
A constituent part of an intracellular organelle, an organized structure of distinctive morphology and function, occurring within the cell. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton but excludes the plasma membrane.
contractile fiber part
Any constituent part of a contractile fiber, a fiber composed of actin, myosin, and associated proteins, found in cells of smooth or striated muscle.
plasma membrane part
Any constituent part of the plasma membrane, the membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins.
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
anchoring junction
A cell junction that mechanically attaches a cell (and its cytoskeleton) to neighboring cells or to the extracellular matrix.
all
NA
extracellular region part
Any constituent part of the extracellular region, the space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers constituent parts of the host cell environment outside an intracellular parasite.
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
organelle part
Any constituent part of an organelle, an organized structure of distinctive morphology and function. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton, but excludes the plasma membrane.
intracellular non-membrane-bounded organelle
Organized structure of distinctive morphology and function, not bounded by a lipid bilayer membrane and occurring within the cell. Includes ribosomes, the cytoskeleton and chromosomes.
intracellular organelle part
A constituent part of an intracellular organelle, an organized structure of distinctive morphology and function, occurring within the cell. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton but excludes the plasma membrane.
extracellular matrix part
Any constituent part of the extracellular matrix, the structure lying external to one or more cells, which provides structural support for cells or tissues; may be completely external to the cell (as in animals) or be part of the cell (as often seen in plants).
intracellular part
Any constituent part of the living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
membrane part
Any constituent part of a membrane, a double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
intracellular organelle
Organized structure of distinctive morphology and function, occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton. Excludes the plasma membrane.
intracellular organelle part
A constituent part of an intracellular organelle, an organized structure of distinctive morphology and function, occurring within the cell. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton but excludes the plasma membrane.
basement membrane
A thin layer of dense material found in various animal tissues interposed between the cells and the adjacent connective tissue. It consists of the basal lamina plus an associated layer of reticulin fibers.
striated muscle thin filament
Filaments formed of actin and associated proteins; attached to Z discs at either end of sarcomeres in myofibrils.
myosin complex
A protein complex, formed of one or more myosin heavy chains plus associated light chains and other proteins, that functions as a molecular motor; uses the energy of ATP hydrolysis to move actin filaments or to move vesicles or other cargo on fixed actin filaments; has magnesium-ATPase activity and binds actin. Myosin classes are distinguished based on sequence features of the motor, or head, domain, but also have distinct tail regions that are believed to bind specific cargoes.
striated muscle thin filament
Filaments formed of actin and associated proteins; attached to Z discs at either end of sarcomeres in myofibrils.
A band
The dark-staining region of a sarcomere, in which myosin thick filaments are present; the center is traversed by the paler H zone, which in turn contains the M line.
I band
A region of a sarcomere that appears as a light band on each side of the Z disc, comprising a region of the sarcomere where thin (actin) filaments are not overlapped by thick (myosin) filaments; contains actin, troponin, and tropomyosin; each sarcomere includes half of an I band at each end.
Z disc
Platelike region of a muscle sarcomere to which the plus ends of actin filaments are attached.
plasma membrane part
Any constituent part of the plasma membrane, the membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins.
cytoplasmic part
Any constituent part of the cytoplasm, all of the contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures.
contractile fiber
Fibers, composed of actin, myosin, and associated proteins, found in cells of smooth or striated muscle.
contractile fiber part
Any constituent part of a contractile fiber, a fiber composed of actin, myosin, and associated proteins, found in cells of smooth or striated muscle.
muscle myosin complex
A filament of myosin found in a muscle cell of any type.
contractile fiber part
Any constituent part of a contractile fiber, a fiber composed of actin, myosin, and associated proteins, found in cells of smooth or striated muscle.
cytoskeletal part
Any constituent part of the cytoskeleton, a cellular scaffolding or skeleton that maintains cell shape, enables some cell motion (using structures such as flagella and cilia), and plays important roles in both intra-cellular transport (e.g. the movement of vesicles and organelles) and cellular division. Includes constituent parts of intermediate filaments, microfilaments, microtubules, and the microtrabecular lattice.
cell-substrate junction
A cell junction that forms a connection between a cell and the extracellular matrix.
sarcomere
The repeating unit of a myofibril in a muscle cell, composed of an array of overlapping thick and thin filaments between two adjacent Z discs.
striated muscle thin filament
Filaments formed of actin and associated proteins; attached to Z discs at either end of sarcomeres in myofibrils.
myosin complex
A protein complex, formed of one or more myosin heavy chains plus associated light chains and other proteins, that functions as a molecular motor; uses the energy of ATP hydrolysis to move actin filaments or to move vesicles or other cargo on fixed actin filaments; has magnesium-ATPase activity and binds actin. Myosin classes are distinguished based on sequence features of the motor, or head, domain, but also have distinct tail regions that are believed to bind specific cargoes.
cell-substrate adherens junction
An adherens junction which connects a cell to the extracellular matrix.

protein binding
Interacting selectively and non-covalently with any protein or protein complex (a complex of two or more proteins that may include other nonprotein molecules).
calcium ion binding
Interacting selectively and non-covalently with calcium ions (Ca2+).
molecular_function
Elemental activities, such as catalysis or binding, describing the actions of a gene product at the molecular level. A given gene product may exhibit one or more molecular functions.
actin binding
Interacting selectively and non-covalently with monomeric or multimeric forms of actin, including actin filaments.
structural molecule activity
The action of a molecule that contributes to the structural integrity of a complex or assembly within or outside a cell.
extracellular matrix structural constituent
The action of a molecule that contributes to the structural integrity of the extracellular matrix.
binding
The selective, non-covalent, often stoichiometric, interaction of a molecule with one or more specific sites on another molecule.
metal ion binding
Interacting selectively and non-covalently with any metal ion.
tropomyosin binding
Interacting selectively and non-covalently with tropomyosin, a protein associated with actin filaments both in cytoplasm and, in association with troponin, in the thin filament of striated muscle.
cytoskeletal protein binding
Interacting selectively and non-covalently with any protein component of any cytoskeleton (actin, microtubule, or intermediate filament cytoskeleton).
structural constituent of muscle
The action of a molecule that contributes to the structural integrity of a muscle fiber.
ion binding
Interacting selectively and non-covalently with ions, charged atoms or groups of atoms.
cation binding
Interacting selectively and non-covalently with cations, charged atoms or groups of atoms with a net positive charge.
all
NA
| Id | Pvalue | ExpCount | Count | Size | Term |
|---|---|---|---|---|---|
| 05414 | 3.008e-05 | 2.399 | 13 | 34 | Dilated cardiomyopathy |
| 05410 | 1.912e-04 | 2.399 | 12 | 34 | Hypertrophic cardiomyopathy (HCM) |
| 04260 | 3.569e-04 | 1.764 | 10 | 25 | Cardiac muscle contraction |
| 05412 | 2.263e-03 | 2.188 | 10 | 31 | Arrhythmogenic right ventricular cardiomyopathy (ARVC) |
ABI3BPABI family, member 3 (NESH) binding protein (ENSG00000154175), score: 0.5 ABLIM1actin binding LIM protein 1 (ENSG00000099204), score: 0.45 ABRAactin-binding Rho activating protein (ENSG00000174429), score: 0.57 ACAP2ArfGAP with coiled-coil, ankyrin repeat and PH domains 2 (ENSG00000114331), score: -0.42 ACTN1actinin, alpha 1 (ENSG00000072110), score: 0.38 ACTN2actinin, alpha 2 (ENSG00000077522), score: 0.51 ADAMTS15ADAM metallopeptidase with thrombospondin type 1 motif, 15 (ENSG00000166106), score: 0.59 ADAMTSL5ADAMTS-like 5 (ENSG00000185761), score: 0.65 ADCY5adenylate cyclase 5 (ENSG00000173175), score: 0.49 ADIPOQadiponectin, C1Q and collagen domain containing (ENSG00000181092), score: 0.78 ADSSadenylosuccinate synthase (ENSG00000035687), score: -0.41 AFAP1L1actin filament associated protein 1-like 1 (ENSG00000157510), score: 0.44 AFTPHaftiphilin (ENSG00000119844), score: -0.47 AIFM2apoptosis-inducing factor, mitochondrion-associated, 2 (ENSG00000042286), score: 0.4 AKAP13A kinase (PRKA) anchor protein 13 (ENSG00000170776), score: 0.48 AMOTL1angiomotin like 1 (ENSG00000166025), score: 0.4 ANGPTL2angiopoietin-like 2 (ENSG00000136859), score: 0.44 ANGPTL7angiopoietin-like 7 (ENSG00000171819), score: 0.77 ANKRD1ankyrin repeat domain 1 (cardiac muscle) (ENSG00000148677), score: 0.64 ANKRD28ankyrin repeat domain 28 (ENSG00000206560), score: -0.49 ANXA1annexin A1 (ENSG00000135046), score: 0.46 AP1G1adaptor-related protein complex 1, gamma 1 subunit (ENSG00000166747), score: -0.47 APOBEC2apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 2 (ENSG00000124701), score: 0.54 APPBP2amyloid beta precursor protein (cytoplasmic tail) binding protein 2 (ENSG00000062725), score: -0.65 ARFRP1ADP-ribosylation factor related protein 1 (ENSG00000101246), score: 0.42 ARHGAP10Rho GTPase activating protein 10 (ENSG00000071205), score: 0.41 ARHGDIGRho GDP dissociation inhibitor (GDI) gamma (ENSG00000167930), score: 0.4 ARHGEF6Rac/Cdc42 guanine nucleotide exchange factor (GEF) 6 (ENSG00000129675), score: 0.4 ARL6IP1ADP-ribosylation factor-like 6 interacting protein 1 (ENSG00000170540), score: -0.44 ARL6IP5ADP-ribosylation-like factor 6 interacting protein 5 (ENSG00000144746), score: 0.38 ART3ADP-ribosyltransferase 3 (ENSG00000156219), score: 0.4 ASB11ankyrin repeat and SOCS box-containing 11 (ENSG00000165192), score: 0.71 ASB15ankyrin repeat and SOCS box-containing 15 (ENSG00000146809), score: 0.52 ASB2ankyrin repeat and SOCS box-containing 2 (ENSG00000100628), score: 0.56 ATF3activating transcription factor 3 (ENSG00000162772), score: 0.41 ATG4CATG4 autophagy related 4 homolog C (S. cerevisiae) (ENSG00000125703), score: -0.39 ATP2A2ATPase, Ca++ transporting, cardiac muscle, slow twitch 2 (ENSG00000174437), score: 0.47 AVL9AVL9 homolog (S. cerevisiase) (ENSG00000105778), score: -0.48 B3GALTLbeta 1,3-galactosyltransferase-like (ENSG00000187676), score: 0.44 B4GALNT3beta-1,4-N-acetyl-galactosaminyl transferase 3 (ENSG00000139044), score: 0.44 BAG3BCL2-associated athanogene 3 (ENSG00000151929), score: 0.43 BMP10bone morphogenetic protein 10 (ENSG00000163217), score: 0.62 BMP7bone morphogenetic protein 7 (ENSG00000101144), score: 0.4 BMXBMX non-receptor tyrosine kinase (ENSG00000102010), score: 0.55 BRF2BRF2, subunit of RNA polymerase III transcription initiation factor, BRF1-like (ENSG00000104221), score: 0.44 C10orf54chromosome 10 open reading frame 54 (ENSG00000107738), score: 0.5 C10orf76chromosome 10 open reading frame 76 (ENSG00000120029), score: 0.38 C14orf49chromosome 14 open reading frame 49 (ENSG00000176438), score: 0.43 C1QTNF2C1q and tumor necrosis factor related protein 2 (ENSG00000145861), score: 0.4 C6orf142chromosome 6 open reading frame 142 (ENSG00000146147), score: 0.51 C7orf58chromosome 7 open reading frame 58 (ENSG00000106034), score: 0.43 C8orf42chromosome 8 open reading frame 42 (ENSG00000180190), score: -0.47 CACNA1Ccalcium channel, voltage-dependent, L type, alpha 1C subunit (ENSG00000151067), score: 0.4 CALHM2calcium homeostasis modulator 2 (ENSG00000138172), score: 0.49 CAPZA2capping protein (actin filament) muscle Z-line, alpha 2 (ENSG00000198898), score: 0.42 CASTcalpastatin (ENSG00000153113), score: 0.44 CCDC3coiled-coil domain containing 3 (ENSG00000151468), score: 0.4 CCDC80coiled-coil domain containing 80 (ENSG00000091986), score: 0.57 CCND2cyclin D2 (ENSG00000118971), score: 0.42 CD151CD151 molecule (Raph blood group) (ENSG00000177697), score: 0.44 CD274CD274 molecule (ENSG00000120217), score: 0.51 CD34CD34 molecule (ENSG00000174059), score: 0.51 CD79BCD79b molecule, immunoglobulin-associated beta (ENSG00000007312), score: 0.43 CDC42EP3CDC42 effector protein (Rho GTPase binding) 3 (ENSG00000163171), score: 0.39 CDCA7Lcell division cycle associated 7-like (ENSG00000164649), score: 0.49 CDH13cadherin 13, H-cadherin (heart) (ENSG00000140945), score: 0.46 CDH2cadherin 2, type 1, N-cadherin (neuronal) (ENSG00000170558), score: 0.44 CDH5cadherin 5, type 2 (vascular endothelium) (ENSG00000179776), score: 0.41 CDS1CDP-diacylglycerol synthase (phosphatidate cytidylyltransferase) 1 (ENSG00000163624), score: -0.41 CHIC2cysteine-rich hydrophobic domain 2 (ENSG00000109220), score: 0.42 CHRNA1cholinergic receptor, nicotinic, alpha 1 (muscle) (ENSG00000138435), score: 0.78 CHRNA10cholinergic receptor, nicotinic, alpha 10 (ENSG00000129749), score: 0.61 CIITAclass II, major histocompatibility complex, transactivator (ENSG00000179583), score: 0.39 CLCN5chloride channel 5 (ENSG00000171365), score: -0.37 CLIC5chloride intracellular channel 5 (ENSG00000112782), score: 0.56 CMYA5cardiomyopathy associated 5 (ENSG00000164309), score: 0.54 COL15A1collagen, type XV, alpha 1 (ENSG00000204291), score: 0.61 COL3A1collagen, type III, alpha 1 (ENSG00000168542), score: 0.46 COL4A2collagen, type IV, alpha 2 (ENSG00000134871), score: 0.46 COL6A2collagen, type VI, alpha 2 (ENSG00000142173), score: 0.55 COL6A3collagen, type VI, alpha 3 (ENSG00000163359), score: 0.45 COMMD8COMM domain containing 8 (ENSG00000169019), score: -0.36 COQ9coenzyme Q9 homolog (S. cerevisiae) (ENSG00000088682), score: 0.47 COX4I1cytochrome c oxidase subunit IV isoform 1 (ENSG00000131143), score: 0.38 CREB3L1cAMP responsive element binding protein 3-like 1 (ENSG00000157613), score: 0.38 CRISPLD2cysteine-rich secretory protein LCCL domain containing 2 (ENSG00000103196), score: 0.48 CRYABcrystallin, alpha B (ENSG00000109846), score: 0.39 CSRP3cysteine and glycine-rich protein 3 (cardiac LIM protein) (ENSG00000129170), score: 0.59 CTRLchymotrypsin-like (ENSG00000141086), score: 0.43 CUL4Acullin 4A (ENSG00000139842), score: 0.4 DCAF6DDB1 and CUL4 associated factor 6 (ENSG00000143164), score: 0.38 DCNdecorin (ENSG00000011465), score: 0.46 DCUN1D2DCN1, defective in cullin neddylation 1, domain containing 2 (S. cerevisiae) (ENSG00000150401), score: 0.49 DDAH1dimethylarginine dimethylaminohydrolase 1 (ENSG00000153904), score: -0.41 DDR2discoidin domain receptor tyrosine kinase 2 (ENSG00000162733), score: 0.44 DICER1dicer 1, ribonuclease type III (ENSG00000100697), score: -0.47 DKK3dickkopf homolog 3 (Xenopus laevis) (ENSG00000050165), score: 0.45 DLK1delta-like 1 homolog (Drosophila) (ENSG00000185559), score: 0.54 DNAJC12DnaJ (Hsp40) homolog, subfamily C, member 12 (ENSG00000108176), score: -0.51 DPTdermatopontin (ENSG00000143196), score: 0.42 DSPdesmoplakin (ENSG00000096696), score: 0.46 DTX4deltex homolog 4 (Drosophila) (ENSG00000110042), score: -0.38 DUSP27dual specificity phosphatase 27 (putative) (ENSG00000198842), score: 0.7 DYRK1Adual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1A (ENSG00000157540), score: 0.5 EEF2Keukaryotic elongation factor-2 kinase (ENSG00000103319), score: 0.45 EHD4EH-domain containing 4 (ENSG00000103966), score: 0.45 EIF3Aeukaryotic translation initiation factor 3, subunit A (ENSG00000107581), score: 0.43 EMILIN2elastin microfibril interfacer 2 (ENSG00000132205), score: 0.53 EMP1epithelial membrane protein 1 (ENSG00000134531), score: 0.41 ESYT2extended synaptotagmin-like protein 2 (ENSG00000117868), score: 0.49 EYA4eyes absent homolog 4 (Drosophila) (ENSG00000112319), score: 0.45 FAM114A1family with sequence similarity 114, member A1 (ENSG00000197712), score: 0.4 FAM129Afamily with sequence similarity 129, member A (ENSG00000135842), score: 0.76 FAM198Bfamily with sequence similarity 198, member B (ENSG00000164125), score: 0.48 FAM46Afamily with sequence similarity 46, member A (ENSG00000112773), score: 0.41 FBLIM1filamin binding LIM protein 1 (ENSG00000162458), score: 0.42 FBLN1fibulin 1 (ENSG00000077942), score: 0.55 FBLN2fibulin 2 (ENSG00000163520), score: 0.62 FBN1fibrillin 1 (ENSG00000166147), score: 0.53 FBXO32F-box protein 32 (ENSG00000156804), score: 0.45 FBXO33F-box protein 33 (ENSG00000165355), score: -0.46 FBXO40F-box protein 40 (ENSG00000163833), score: 0.63 FGF12fibroblast growth factor 12 (ENSG00000114279), score: 0.4 FHL2four and a half LIM domains 2 (ENSG00000115641), score: 0.49 FHOD1formin homology 2 domain containing 1 (ENSG00000135723), score: 0.44 FHOD3formin homology 2 domain containing 3 (ENSG00000134775), score: 0.53 FILIP1filamin A interacting protein 1 (ENSG00000118407), score: 0.49 FITM2fat storage-inducing transmembrane protein 2 (ENSG00000197296), score: 0.73 FKBP10FK506 binding protein 10, 65 kDa (ENSG00000141756), score: 0.45 FLIIflightless I homolog (Drosophila) (ENSG00000177731), score: 0.7 FRZBfrizzled-related protein (ENSG00000162998), score: 0.58 FSD2fibronectin type III and SPRY domain containing 2 (ENSG00000186628), score: 0.49 FUT11fucosyltransferase 11 (alpha (1,3) fucosyltransferase) (ENSG00000196968), score: 0.42 FYCO1FYVE and coiled-coil domain containing 1 (ENSG00000163820), score: 0.52 GALNT5UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 5 (GalNAc-T5) (ENSG00000136542), score: 0.5 GAS6growth arrest-specific 6 (ENSG00000183087), score: 0.51 GATA4GATA binding protein 4 (ENSG00000136574), score: 0.44 GATA6GATA binding protein 6 (ENSG00000141448), score: 0.48 GCOM1GRINL1A complex locus (ENSG00000137878), score: 0.43 GEMGTP binding protein overexpressed in skeletal muscle (ENSG00000164949), score: 0.54 GGHgamma-glutamyl hydrolase (conjugase, folylpolygammaglutamyl hydrolase) (ENSG00000137563), score: -0.37 GJA3gap junction protein, alpha 3, 46kDa (ENSG00000121743), score: 0.62 GJA5gap junction protein, alpha 5, 40kDa (ENSG00000143140), score: 0.49 GJB1gap junction protein, beta 1, 32kDa (ENSG00000169562), score: -0.39 GLDCglycine dehydrogenase (decarboxylating) (ENSG00000178445), score: -0.4 GLP1Rglucagon-like peptide 1 receptor (ENSG00000112164), score: 0.51 GLT1D1glycosyltransferase 1 domain containing 1 (ENSG00000151948), score: -0.4 GLT8D2glycosyltransferase 8 domain containing 2 (ENSG00000120820), score: 0.44 GMPRguanosine monophosphate reductase (ENSG00000137198), score: 0.47 GPNMBglycoprotein (transmembrane) nmb (ENSG00000136235), score: 0.44 GPR124G protein-coupled receptor 124 (ENSG00000020181), score: 0.44 GPR133G protein-coupled receptor 133 (ENSG00000111452), score: 0.53 GPR183G protein-coupled receptor 183 (ENSG00000169508), score: 0.38 GPR20G protein-coupled receptor 20 (ENSG00000204882), score: 0.5 GPX8glutathione peroxidase 8 (putative) (ENSG00000164294), score: 0.38 GRAMD1CGRAM domain containing 1C (ENSG00000178075), score: -0.38 GRIP2glutamate receptor interacting protein 2 (ENSG00000144596), score: 0.51 GRK5G protein-coupled receptor kinase 5 (ENSG00000198873), score: 0.45 GRXCR2glutaredoxin, cysteine rich 2 (ENSG00000204928), score: 0.38 HEATR2HEAT repeat containing 2 (ENSG00000164818), score: 0.44 HEY2hairy/enhancer-of-split related with YRPW motif 2 (ENSG00000135547), score: 0.51 HEYLhairy/enhancer-of-split related with YRPW motif-like (ENSG00000163909), score: 0.5 HHATLhedgehog acyltransferase-like (ENSG00000010282), score: 0.51 HMCN1hemicentin 1 (ENSG00000143341), score: 0.38 HOOK1hook homolog 1 (Drosophila) (ENSG00000134709), score: -0.41 HRH2histamine receptor H2 (ENSG00000113749), score: 0.42 HSPG2heparan sulfate proteoglycan 2 (ENSG00000142798), score: 0.47 IGF2Rinsulin-like growth factor 2 receptor (ENSG00000197081), score: 0.45 IL17Binterleukin 17B (ENSG00000127743), score: 0.43 IL20RAinterleukin 20 receptor, alpha (ENSG00000016402), score: 0.59 INHBAinhibin, beta A (ENSG00000122641), score: 0.43 ITGA8integrin, alpha 8 (ENSG00000077943), score: 0.41 ITPRIPinositol 1,4,5-triphosphate receptor interacting protein (ENSG00000148841), score: 0.42 KAZALD1Kazal-type serine peptidase inhibitor domain 1 (ENSG00000107821), score: 0.49 KBTBD10kelch repeat and BTB (POZ) domain containing 10 (ENSG00000239474), score: 0.6 KBTBD5kelch repeat and BTB (POZ) domain containing 5 (ENSG00000157119), score: 0.67 KCNA5potassium voltage-gated channel, shaker-related subfamily, member 5 (ENSG00000130037), score: 0.71 KCNJ2potassium inwardly-rectifying channel, subfamily J, member 2 (ENSG00000123700), score: 0.46 KCNJ5potassium inwardly-rectifying channel, subfamily J, member 5 (ENSG00000120457), score: 0.54 KCNJ8potassium inwardly-rectifying channel, subfamily J, member 8 (ENSG00000121361), score: 0.4 KCNMB1potassium large conductance calcium-activated channel, subfamily M, beta member 1 (ENSG00000145936), score: 0.66 KCNMB2potassium large conductance calcium-activated channel, subfamily M, beta member 2 (ENSG00000197584), score: 0.38 KCNQ1potassium voltage-gated channel, KQT-like subfamily, member 1 (ENSG00000053918), score: 0.4 KIAA1598KIAA1598 (ENSG00000187164), score: -0.47 KIAA1958KIAA1958 (ENSG00000165185), score: -0.36 KIF13Akinesin family member 13A (ENSG00000137177), score: 0.48 KLHL21kelch-like 21 (Drosophila) (ENSG00000162413), score: 0.46 KLHL31kelch-like 31 (Drosophila) (ENSG00000124743), score: 0.66 KLHL38kelch-like 38 (Drosophila) (ENSG00000175946), score: 0.62 KLHL7kelch-like 7 (Drosophila) (ENSG00000122550), score: 0.39 LAMA4laminin, alpha 4 (ENSG00000112769), score: 0.6 LAMB2laminin, beta 2 (laminin S) (ENSG00000172037), score: 0.55 LAPTM4Blysosomal protein transmembrane 4 beta (ENSG00000104341), score: 0.48 LARP4BLa ribonucleoprotein domain family, member 4B (ENSG00000107929), score: 0.48 LBHlimb bud and heart development homolog (mouse) (ENSG00000213626), score: 0.41 LDB3LIM domain binding 3 (ENSG00000122367), score: 0.6 LIMS2LIM and senescent cell antigen-like domains 2 (ENSG00000072163), score: 0.59 LMOD2leiomodin 2 (cardiac) (ENSG00000170807), score: 0.64 LMOD3leiomodin 3 (fetal) (ENSG00000163380), score: 0.51 LOC100291405similar to protein tyrosine phosphatase-like A domain containing 1 (ENSG00000074696), score: -0.39 LOC100291671similar to SH3-binding domain and glutamic acid-rich protein (ENSG00000185437), score: 0.39 LRIG1leucine-rich repeats and immunoglobulin-like domains 1 (ENSG00000144749), score: 0.41 LRRC10leucine rich repeat containing 10 (ENSG00000198812), score: 0.56 LRRC14Bleucine rich repeat containing 14B (ENSG00000185028), score: 0.4 LRRC17leucine rich repeat containing 17 (ENSG00000128606), score: 0.42 LRRC2leucine rich repeat containing 2 (ENSG00000163827), score: 0.51 LRRC39leucine rich repeat containing 39 (ENSG00000122477), score: 0.62 LTBP1latent transforming growth factor beta binding protein 1 (ENSG00000049323), score: 0.63 LUMlumican (ENSG00000139329), score: 0.48 MAFKv-maf musculoaponeurotic fibrosarcoma oncogene homolog K (avian) (ENSG00000198517), score: 0.45 MAMDC2MAM domain containing 2 (ENSG00000165072), score: 0.6 MAN1A2mannosidase, alpha, class 1A, member 2 (ENSG00000198162), score: -0.4 MAP3K2mitogen-activated protein kinase kinase kinase 2 (ENSG00000169967), score: -0.37 MAP7microtubule-associated protein 7 (ENSG00000135525), score: -0.46 MAPKAPK2mitogen-activated protein kinase-activated protein kinase 2 (ENSG00000162889), score: 0.5 MAPKAPK3mitogen-activated protein kinase-activated protein kinase 3 (ENSG00000114738), score: 0.42 MATN2matrilin 2 (ENSG00000132561), score: 0.48 MCCmutated in colorectal cancers (ENSG00000171444), score: 0.42 ME2malic enzyme 2, NAD(+)-dependent, mitochondrial (ENSG00000082212), score: 0.4 MEOX2mesenchyme homeobox 2 (ENSG00000106511), score: 0.44 MFN2mitofusin 2 (ENSG00000116688), score: 0.45 MLPHmelanophilin (ENSG00000115648), score: 0.51 MLXIPMLX interacting protein (ENSG00000175727), score: 0.39 MMP2matrix metallopeptidase 2 (gelatinase A, 72kDa gelatinase, 72kDa type IV collagenase) (ENSG00000087245), score: 0.51 MRPS25mitochondrial ribosomal protein S25 (ENSG00000131368), score: 0.38 MRPS9mitochondrial ribosomal protein S9 (ENSG00000135972), score: 0.46 MST1Rmacrophage stimulating 1 receptor (c-met-related tyrosine kinase) (ENSG00000164078), score: 0.4 MTIF2mitochondrial translational initiation factor 2 (ENSG00000085760), score: 0.38 MURCmuscle-related coiled-coil protein (ENSG00000170681), score: 0.47 MYBPC3myosin binding protein C, cardiac (ENSG00000134571), score: 0.61 MYH11myosin, heavy chain 11, smooth muscle (ENSG00000133392), score: 0.6 MYL1myosin, light chain 1, alkali; skeletal, fast (ENSG00000168530), score: 0.51 MYL3myosin, light chain 3, alkali; ventricular, skeletal, slow (ENSG00000160808), score: 0.52 MYL4myosin, light chain 4, alkali; atrial, embryonic (ENSG00000198336), score: 0.87 MYLK3myosin light chain kinase 3 (ENSG00000140795), score: 0.52 MYO18Amyosin XVIIIA (ENSG00000196535), score: 0.39 MYO1Cmyosin IC (ENSG00000197879), score: 0.41 MYO6myosin VI (ENSG00000196586), score: -0.42 MYOCmyocilin, trabecular meshwork inducible glucocorticoid response (ENSG00000034971), score: 0.64 MYOCDmyocardin (ENSG00000141052), score: 0.6 MYOM2myomesin (M-protein) 2, 165kDa (ENSG00000036448), score: 0.59 MYOM3myomesin family, member 3 (ENSG00000142661), score: 0.72 MYOZ1myozenin 1 (ENSG00000177791), score: 0.81 MYOZ2myozenin 2 (ENSG00000172399), score: 0.59 MYPNmyopalladin (ENSG00000138347), score: 0.64 NAA25N(alpha)-acetyltransferase 25, NatB auxiliary subunit (ENSG00000111300), score: -0.38 NDE1nudE nuclear distribution gene E homolog 1 (A. nidulans) (ENSG00000072864), score: 0.45 NDUFB10NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 10, 22kDa (ENSG00000140990), score: 0.45 NEBLnebulette (ENSG00000078114), score: 0.42 NEDD9neural precursor cell expressed, developmentally down-regulated 9 (ENSG00000111859), score: 0.41 NFATC1nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 1 (ENSG00000131196), score: 0.43 NGEFneuronal guanine nucleotide exchange factor (ENSG00000066248), score: -0.4 NID1nidogen 1 (ENSG00000116962), score: 0.4 NLRC3NLR family, CARD domain containing 3 (ENSG00000167984), score: 0.43 NLRX1NLR family member X1 (ENSG00000160703), score: 0.49 NOVnephroblastoma overexpressed gene (ENSG00000136999), score: 0.44 NPR3natriuretic peptide receptor C/guanylate cyclase C (atrionatriuretic peptide receptor C) (ENSG00000113389), score: 0.42 NRAPnebulin-related anchoring protein (ENSG00000197893), score: 0.56 NRTNneurturin (ENSG00000171119), score: 0.43 NTN4netrin 4 (ENSG00000074527), score: 0.47 OBFC1oligonucleotide/oligosaccharide-binding fold containing 1 (ENSG00000107960), score: 0.47 OGDHoxoglutarate (alpha-ketoglutarate) dehydrogenase (lipoamide) (ENSG00000105953), score: 0.41 OLFML2Aolfactomedin-like 2A (ENSG00000185585), score: 0.47 OTUD1OTU domain containing 1 (ENSG00000165312), score: 0.4 P2RY2purinergic receptor P2Y, G-protein coupled, 2 (ENSG00000175591), score: 0.41 PAMpeptidylglycine alpha-amidating monooxygenase (ENSG00000145730), score: 0.61 PARD6Apar-6 partitioning defective 6 homolog alpha (C. elegans) (ENSG00000102981), score: -0.41 PCDH18protocadherin 18 (ENSG00000189184), score: 0.4 PCYT1Aphosphate cytidylyltransferase 1, choline, alpha (ENSG00000161217), score: 0.39 PDE3Aphosphodiesterase 3A, cGMP-inhibited (ENSG00000172572), score: 0.44 PDE7Aphosphodiesterase 7A (ENSG00000205268), score: 0.4 PDGFRLplatelet-derived growth factor receptor-like (ENSG00000104213), score: 0.44 PDLIM5PDZ and LIM domain 5 (ENSG00000163110), score: 0.51 PDZRN3PDZ domain containing ring finger 3 (ENSG00000121440), score: 0.58 PHF10PHD finger protein 10 (ENSG00000130024), score: 0.45 PIK3C2Aphosphoinositide-3-kinase, class 2, alpha polypeptide (ENSG00000011405), score: -0.41 PLBD1phospholipase B domain containing 1 (ENSG00000121316), score: 0.46 PLEKHA2pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 2 (ENSG00000169499), score: 0.39 PLIN1perilipin 1 (ENSG00000166819), score: 1 PLNphospholamban (ENSG00000198523), score: 0.52 PODNpodocan (ENSG00000174348), score: 0.44 PPICpeptidylprolyl isomerase C (cyclophilin C) (ENSG00000168938), score: 0.38 PPP1R1Cprotein phosphatase 1, regulatory (inhibitor) subunit 1C (ENSG00000150722), score: 0.4 PPP1R3Aprotein phosphatase 1, regulatory (inhibitor) subunit 3A (ENSG00000154415), score: 0.59 PRELPproline/arginine-rich end leucine-rich repeat protein (ENSG00000188783), score: 0.55 PRKAG2protein kinase, AMP-activated, gamma 2 non-catalytic subunit (ENSG00000106617), score: 0.38 PSTPIP1proline-serine-threonine phosphatase interacting protein 1 (ENSG00000140368), score: 0.38 PTCD2pentatricopeptide repeat domain 2 (ENSG00000049883), score: 0.4 PTDSS1phosphatidylserine synthase 1 (ENSG00000156471), score: 0.46 PTGFRNprostaglandin F2 receptor negative regulator (ENSG00000134247), score: 0.56 PTRFpolymerase I and transcript release factor (ENSG00000177469), score: 0.53 QARSglutaminyl-tRNA synthetase (ENSG00000172053), score: 0.43 RAB11FIP4RAB11 family interacting protein 4 (class II) (ENSG00000131242), score: -0.36 RAMP3receptor (G protein-coupled) activity modifying protein 3 (ENSG00000122679), score: 0.38 RASL12RAS-like, family 12 (ENSG00000103710), score: 0.45 RBM20RNA binding motif protein 20 (ENSG00000203867), score: 0.52 RBM24RNA binding motif protein 24 (ENSG00000112183), score: 0.54 RBPMS2RNA binding protein with multiple splicing 2 (ENSG00000166831), score: 0.45 REREarginine-glutamic acid dipeptide (RE) repeats (ENSG00000142599), score: 0.43 RILPL1Rab interacting lysosomal protein-like 1 (ENSG00000188026), score: 0.44 ROR1receptor tyrosine kinase-like orphan receptor 1 (ENSG00000185483), score: 0.54 RORARAR-related orphan receptor A (ENSG00000069667), score: -0.36 RPL3Lribosomal protein L3-like (ENSG00000140986), score: 0.5 RRADRas-related associated with diabetes (ENSG00000166592), score: 0.55 RSPO1R-spondin homolog (Xenopus laevis) (ENSG00000169218), score: 0.67 RYR2ryanodine receptor 2 (cardiac) (ENSG00000198626), score: 0.45 SCLT1sodium channel and clathrin linker 1 (ENSG00000151466), score: 0.5 SEMA3Csema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3C (ENSG00000075223), score: 0.38 SERINC4serine incorporator 4 (ENSG00000184716), score: 0.51 SETDB2SET domain, bifurcated 2 (ENSG00000136169), score: 0.58 SFRP2secreted frizzled-related protein 2 (ENSG00000145423), score: 0.42 SFRP4secreted frizzled-related protein 4 (ENSG00000106483), score: 0.55 SGCGsarcoglycan, gamma (35kDa dystrophin-associated glycoprotein) (ENSG00000102683), score: 0.59 SGPL1sphingosine-1-phosphate lyase 1 (ENSG00000166224), score: -0.37 SH3RF2SH3 domain containing ring finger 2 (ENSG00000156463), score: 0.43 SIPA1L1signal-induced proliferation-associated 1 like 1 (ENSG00000197555), score: -0.42 SLC16A1solute carrier family 16, member 1 (monocarboxylic acid transporter 1) (ENSG00000155380), score: 0.43 SLC25A17solute carrier family 25 (mitochondrial carrier; peroxisomal membrane protein, 34kDa), member 17 (ENSG00000100372), score: -0.39 SLC25A33solute carrier family 25, member 33 (ENSG00000171612), score: -0.39 SLC26A9solute carrier family 26, member 9 (ENSG00000174502), score: 0.41 SLC31A2solute carrier family 31 (copper transporters), member 2 (ENSG00000136867), score: -0.42 SLC41A1solute carrier family 41, member 1 (ENSG00000133065), score: 0.62 SLC41A2solute carrier family 41, member 2 (ENSG00000136052), score: -0.38 SLC7A6solute carrier family 7 (cationic amino acid transporter, y+ system), member 6 (ENSG00000103064), score: 0.41 SLC8A1solute carrier family 8 (sodium/calcium exchanger), member 1 (ENSG00000183023), score: 0.45 SMPXsmall muscle protein, X-linked (ENSG00000091482), score: 0.62 SMYD1SET and MYND domain containing 1 (ENSG00000115593), score: 0.54 SMYD2SET and MYND domain containing 2 (ENSG00000143499), score: 0.42 SNX10sorting nexin 10 (ENSG00000086300), score: -0.41 SNX14sorting nexin 14 (ENSG00000135317), score: -0.41 SNX30sorting nexin family member 30 (ENSG00000148158), score: -0.39 SORBS1sorbin and SH3 domain containing 1 (ENSG00000095637), score: 0.48 SPATS2spermatogenesis associated, serine-rich 2 (ENSG00000123352), score: -0.44 STK17Bserine/threonine kinase 17b (ENSG00000081320), score: -0.45 STX11syntaxin 11 (ENSG00000135604), score: 0.54 SYNMsynemin, intermediate filament protein (ENSG00000182253), score: 0.5 SYNPO2synaptopodin 2 (ENSG00000172403), score: 0.51 TACC2transforming, acidic coiled-coil containing protein 2 (ENSG00000138162), score: 0.41 TANC1tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 1 (ENSG00000115183), score: 0.5 TBC1D30TBC1 domain family, member 30 (ENSG00000111490), score: -0.38 TBX18T-box 18 (ENSG00000112837), score: 0.55 TBX20T-box 20 (ENSG00000164532), score: 0.62 TBX5T-box 5 (ENSG00000089225), score: 0.68 TEAD3TEA domain family member 3 (ENSG00000007866), score: 0.38 TECRLtrans-2,3-enoyl-CoA reductase-like (ENSG00000205678), score: 0.7 TGFAtransforming growth factor, alpha (ENSG00000163235), score: -0.39 TGM2transglutaminase 2 (C polypeptide, protein-glutamine-gamma-glutamyltransferase) (ENSG00000198959), score: 0.39 TIMP3TIMP metallopeptidase inhibitor 3 (ENSG00000100234), score: 0.38 TLCD1TLC domain containing 1 (ENSG00000160606), score: -0.49 TLK1tousled-like kinase 1 (ENSG00000198586), score: -0.37 TLN1talin 1 (ENSG00000137076), score: 0.55 TMEM164transmembrane protein 164 (ENSG00000157600), score: 0.39 TMEM182transmembrane protein 182 (ENSG00000170417), score: 0.61 TMEM43transmembrane protein 43 (ENSG00000170876), score: 0.54 TMEM57transmembrane protein 57 (ENSG00000204178), score: -0.41 TMOD1tropomodulin 1 (ENSG00000136842), score: 0.41 TMTC3transmembrane and tetratricopeptide repeat containing 3 (ENSG00000139324), score: -0.4 TNFRSF19tumor necrosis factor receptor superfamily, member 19 (ENSG00000127863), score: 0.47 TNNC1troponin C type 1 (slow) (ENSG00000114854), score: 0.55 TNNT2troponin T type 2 (cardiac) (ENSG00000118194), score: 0.61 TPD52tumor protein D52 (ENSG00000076554), score: -0.42 TPM1tropomyosin 1 (alpha) (ENSG00000140416), score: 0.55 TRIM29tripartite motif-containing 29 (ENSG00000137699), score: 0.45 TRIM55tripartite motif-containing 55 (ENSG00000147573), score: 0.49 TRIM63tripartite motif-containing 63 (ENSG00000158022), score: 0.53 TSPAN18tetraspanin 18 (ENSG00000157570), score: 0.43 TTC39Btetratricopeptide repeat domain 39B (ENSG00000155158), score: -0.55 TUBB6tubulin, beta 6 (ENSG00000176014), score: 0.56 TXLNBtaxilin beta (ENSG00000164440), score: 0.53 UBE2G2ubiquitin-conjugating enzyme E2G 2 (UBC7 homolog, yeast) (ENSG00000184787), score: 0.43 UHRF1BP1LUHRF1 binding protein 1-like (ENSG00000111647), score: -0.45 UQCRC2ubiquinol-cytochrome c reductase core protein II (ENSG00000140740), score: 0.38 USP28ubiquitin specific peptidase 28 (ENSG00000048028), score: 0.45 VIPR2vasoactive intestinal peptide receptor 2 (ENSG00000106018), score: 0.7 VLDLRvery low density lipoprotein receptor (ENSG00000147852), score: 0.5 VPS41vacuolar protein sorting 41 homolog (S. cerevisiae) (ENSG00000006715), score: -0.39 WDR1WD repeat domain 1 (ENSG00000071127), score: 0.46 WISP2WNT1 inducible signaling pathway protein 2 (ENSG00000064205), score: 0.65 WNT11wingless-type MMTV integration site family, member 11 (ENSG00000085741), score: 0.42 WTIPWilms tumor 1 interacting protein (ENSG00000142279), score: 0.4 XRCC3X-ray repair complementing defective repair in Chinese hamster cells 3 (ENSG00000126215), score: 0.5 ZAKsterile alpha motif and leucine zipper containing kinase AZK (ENSG00000091436), score: 0.54 ZBTB33zinc finger and BTB domain containing 33 (ENSG00000177485), score: -0.37 ZNF330zinc finger protein 330 (ENSG00000109445), score: 0.44
| Id | species | tissue | sex | individual |
|---|---|---|---|---|
| mml_ht_m_ca1 | mml | ht | m | _ |
| hsa_ht_m1_ca1 | hsa | ht | m | 1 |
| mml_ht_f_ca1 | mml | ht | f | _ |