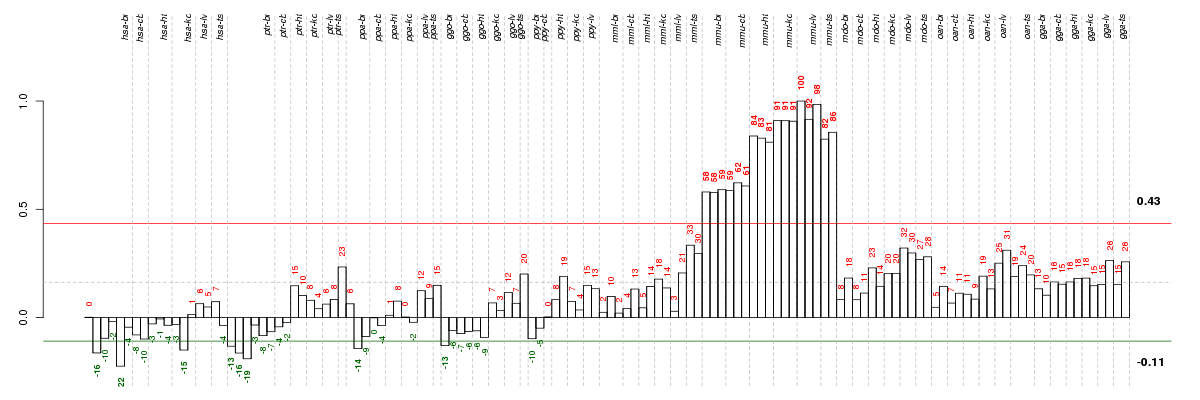

Under-expression is coded with green,
over-expression with red color.

metabolic process
The chemical reactions and pathways, including anabolism and catabolism, by which living organisms transform chemical substances. Metabolic processes typically transform small molecules, but also include macromolecular processes such as DNA repair and replication, and protein synthesis and degradation.
proteolysis
The chemical reactions and pathways resulting in the breakdown of a protein by the destruction of the native, active configuration, with the hydrolysis of peptide bonds.
proteolysis involved in cellular protein catabolic process
The hydrolysis of a peptide bond or bonds within a protein as part of the chemical reactions and pathways resulting in the breakdown of a protein by individual cells.
biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
catabolic process
The chemical reactions and pathways resulting in the breakdown of substances, including the breakdown of carbon compounds with the liberation of energy for use by the cell or organism.
macromolecule catabolic process
The chemical reactions and pathways resulting in the breakdown of a macromolecule, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass.
cellular process
Any process that is carried out at the cellular level, but not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
protein metabolic process
The chemical reactions and pathways involving a specific protein, rather than of proteins in general. Includes protein modification.
protein catabolic process
The chemical reactions and pathways resulting in the breakdown of a protein by the destruction of the native, active configuration, with or without the hydrolysis of peptide bonds.
macromolecule metabolic process
The chemical reactions and pathways involving macromolecules, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass.
cellular metabolic process
The chemical reactions and pathways by which individual cells transform chemical substances.
primary metabolic process
The chemical reactions and pathways involving those compounds which are formed as a part of the normal anabolic and catabolic processes. These processes take place in most, if not all, cells of the organism.
cellular catabolic process
The chemical reactions and pathways resulting in the breakdown of substances, carried out by individual cells.
cellular protein catabolic process
The chemical reactions and pathways resulting in the breakdown of a protein by individual cells.
cellular macromolecule metabolic process
The chemical reactions and pathways involving macromolecules, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass, as carried out by individual cells.
cellular macromolecule catabolic process
The chemical reactions and pathways resulting in the breakdown of a macromolecule, any large molecule including proteins, nucleic acids and carbohydrates, as carried out by individual cells.
cellular protein metabolic process
The chemical reactions and pathways involving a specific protein, rather than of proteins in general, occurring at the level of an individual cell. Includes protein modification.
all
NA
cellular metabolic process
The chemical reactions and pathways by which individual cells transform chemical substances.
macromolecule catabolic process
The chemical reactions and pathways resulting in the breakdown of a macromolecule, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass.
cellular catabolic process
The chemical reactions and pathways resulting in the breakdown of substances, carried out by individual cells.
cellular macromolecule metabolic process
The chemical reactions and pathways involving macromolecules, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass, as carried out by individual cells.
protein metabolic process
The chemical reactions and pathways involving a specific protein, rather than of proteins in general. Includes protein modification.
cellular macromolecule catabolic process
The chemical reactions and pathways resulting in the breakdown of a macromolecule, any large molecule including proteins, nucleic acids and carbohydrates, as carried out by individual cells.
protein catabolic process
The chemical reactions and pathways resulting in the breakdown of a protein by the destruction of the native, active configuration, with or without the hydrolysis of peptide bonds.
cellular macromolecule catabolic process
The chemical reactions and pathways resulting in the breakdown of a macromolecule, any large molecule including proteins, nucleic acids and carbohydrates, as carried out by individual cells.
cellular protein metabolic process
The chemical reactions and pathways involving a specific protein, rather than of proteins in general, occurring at the level of an individual cell. Includes protein modification.
cellular protein catabolic process
The chemical reactions and pathways resulting in the breakdown of a protein by individual cells.
cellular protein catabolic process
The chemical reactions and pathways resulting in the breakdown of a protein by individual cells.
proteolysis involved in cellular protein catabolic process
The hydrolysis of a peptide bond or bonds within a protein as part of the chemical reactions and pathways resulting in the breakdown of a protein by individual cells.

intracellular
The living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
membrane
Double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
integral to membrane
Penetrating at least one phospholipid bilayer of a membrane. May also refer to the state of being buried in the bilayer with no exposure outside the bilayer. When used to describe a protein, indicates that all or part of the peptide sequence is embedded in the membrane.
peroxisome
A small, membrane-bounded organelle that uses dioxygen (O2) to oxidize organic molecules; contains some enzymes that produce and others that degrade hydrogen peroxide (H2O2).
peroxisomal membrane
The lipid bilayer surrounding a peroxisome.
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
cell
The basic structural and functional unit of all organisms. Includes the plasma membrane and any external encapsulating structures such as the cell wall and cell envelope.
cytoplasm
All of the contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures.
integral to peroxisomal membrane
Penetrating at least one phospholipid bilayer of a peroxisomal membrane. May also refer to the state of being buried in the bilayer with no exposure outside the bilayer.
peroxisomal part
Any constituent part of a peroxisome, a small, membrane-bounded organelle that uses dioxygen (O2) to oxidize organic molecules; contains some enzymes that produce and others that degrade hydrogen peroxide (H2O2).
organelle membrane
The lipid bilayer surrounding an organelle.
intrinsic to membrane
Located in a membrane such that some covalently attached portion of the gene product, for example part of a peptide sequence or some other covalently attached moiety such as a GPI anchor, spans or is embedded in one or both leaflets of the membrane.
intrinsic to peroxisomal membrane
Located in the peroxisomal membrane such that some covalently attached portion of the gene product, for example part of a peptide sequence or some other covalently attached moiety such as a GPI anchor, spans or is embedded in one or both leaflets of the membrane.
intrinsic to organelle membrane
Located in an organelle membrane such that some covalently attached portion of the gene product, for example part of a peptide sequence or some other covalently attached moiety such as a GPI anchor, spans or is embedded in one or both leaflets of the membrane.
integral to organelle membrane
Penetrating at least one phospholipid bilayer of an organelle membrane. May also refer to the state of being buried in the bilayer with no exposure outside the bilayer.
microbody membrane
The lipid bilayer surrounding a microbody.
membrane-enclosed lumen
The enclosed volume within a sealed membrane or between two sealed membranes. Encompasses the volume enclosed by the membranes of a particular organelle, e.g. endoplasmic reticulum lumen, or the space between the two lipid bilayers of a double membrane surrounding an organelle, e.g. nuclear envelope lumen.
microbody
Cytoplasmic organelles, spherical or oval in shape, that are bounded by a single membrane and contain oxidative enzymes, especially those utilizing hydrogen peroxide (H2O2).
organelle
Organized structure of distinctive morphology and function. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton, and prokaryotic structures such as anammoxosomes and pirellulosomes. Excludes the plasma membrane.
membrane-bounded organelle
Organized structure of distinctive morphology and function, bounded by a single or double lipid bilayer membrane. Includes the nucleus, mitochondria, plastids, vacuoles, and vesicles. Excludes the plasma membrane.
intracellular organelle
Organized structure of distinctive morphology and function, occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton. Excludes the plasma membrane.
intracellular membrane-bounded organelle
Organized structure of distinctive morphology and function, bounded by a single or double lipid bilayer membrane and occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, and vesicles. Excludes the plasma membrane.
organelle lumen
The internal volume enclosed by the membranes of a particular organelle; includes the volume enclosed by a single organelle membrane, e.g. endoplasmic reticulum lumen, or the volume enclosed by the innermost of the two lipid bilayers of an organelle envelope, e.g. nuclear lumen.
organelle part
Any constituent part of an organelle, an organized structure of distinctive morphology and function. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton, but excludes the plasma membrane.
intracellular part
Any constituent part of the living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
membrane part
Any constituent part of a membrane, a double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
microbody part
Any constituent part of a microbody, a cytoplasmic organelle, spherical or oval in shape, that is bounded by a single membrane and contains oxidative enzymes, especially those utilizing hydrogen peroxide (H2O2).
cytoplasmic part
Any constituent part of the cytoplasm, all of the contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures.
intracellular organelle part
A constituent part of an intracellular organelle, an organized structure of distinctive morphology and function, occurring within the cell. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton but excludes the plasma membrane.
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
intracellular organelle lumen
An organelle lumen that is part of an intracellular organelle.
all
NA
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
organelle part
Any constituent part of an organelle, an organized structure of distinctive morphology and function. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton, but excludes the plasma membrane.
organelle lumen
The internal volume enclosed by the membranes of a particular organelle; includes the volume enclosed by a single organelle membrane, e.g. endoplasmic reticulum lumen, or the volume enclosed by the innermost of the two lipid bilayers of an organelle envelope, e.g. nuclear lumen.
organelle membrane
The lipid bilayer surrounding an organelle.
intracellular membrane-bounded organelle
Organized structure of distinctive morphology and function, bounded by a single or double lipid bilayer membrane and occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, and vesicles. Excludes the plasma membrane.
intracellular organelle part
A constituent part of an intracellular organelle, an organized structure of distinctive morphology and function, occurring within the cell. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton but excludes the plasma membrane.
intrinsic to organelle membrane
Located in an organelle membrane such that some covalently attached portion of the gene product, for example part of a peptide sequence or some other covalently attached moiety such as a GPI anchor, spans or is embedded in one or both leaflets of the membrane.
intracellular organelle lumen
An organelle lumen that is part of an intracellular organelle.
intracellular part
Any constituent part of the living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
organelle membrane
The lipid bilayer surrounding an organelle.
membrane part
Any constituent part of a membrane, a double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
intracellular organelle
Organized structure of distinctive morphology and function, occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton. Excludes the plasma membrane.
intracellular organelle part
A constituent part of an intracellular organelle, an organized structure of distinctive morphology and function, occurring within the cell. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton but excludes the plasma membrane.
cytoplasmic part
Any constituent part of the cytoplasm, all of the contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures.
microbody
Cytoplasmic organelles, spherical or oval in shape, that are bounded by a single membrane and contain oxidative enzymes, especially those utilizing hydrogen peroxide (H2O2).
microbody part
Any constituent part of a microbody, a cytoplasmic organelle, spherical or oval in shape, that is bounded by a single membrane and contains oxidative enzymes, especially those utilizing hydrogen peroxide (H2O2).
intrinsic to organelle membrane
Located in an organelle membrane such that some covalently attached portion of the gene product, for example part of a peptide sequence or some other covalently attached moiety such as a GPI anchor, spans or is embedded in one or both leaflets of the membrane.
microbody membrane
The lipid bilayer surrounding a microbody.
microbody part
Any constituent part of a microbody, a cytoplasmic organelle, spherical or oval in shape, that is bounded by a single membrane and contains oxidative enzymes, especially those utilizing hydrogen peroxide (H2O2).
integral to peroxisomal membrane
Penetrating at least one phospholipid bilayer of a peroxisomal membrane. May also refer to the state of being buried in the bilayer with no exposure outside the bilayer.
intrinsic to peroxisomal membrane
Located in the peroxisomal membrane such that some covalently attached portion of the gene product, for example part of a peptide sequence or some other covalently attached moiety such as a GPI anchor, spans or is embedded in one or both leaflets of the membrane.
peroxisomal membrane
The lipid bilayer surrounding a peroxisome.
intrinsic to peroxisomal membrane
Located in the peroxisomal membrane such that some covalently attached portion of the gene product, for example part of a peptide sequence or some other covalently attached moiety such as a GPI anchor, spans or is embedded in one or both leaflets of the membrane.
integral to organelle membrane
Penetrating at least one phospholipid bilayer of an organelle membrane. May also refer to the state of being buried in the bilayer with no exposure outside the bilayer.
peroxisomal part
Any constituent part of a peroxisome, a small, membrane-bounded organelle that uses dioxygen (O2) to oxidize organic molecules; contains some enzymes that produce and others that degrade hydrogen peroxide (H2O2).

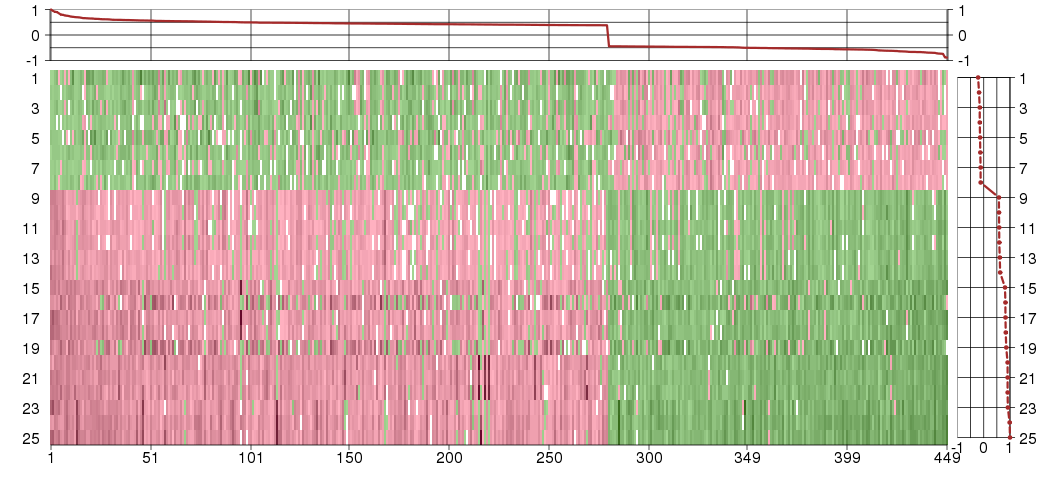
A4GNTalpha-1,4-N-acetylglucosaminyltransferase (ENSG00000118017), score: 0.41 ABCD3ATP-binding cassette, sub-family D (ALD), member 3 (ENSG00000117528), score: 0.51 ABCE1ATP-binding cassette, sub-family E (OABP), member 1 (ENSG00000164163), score: 0.45 ABI2abl-interactor 2 (ENSG00000138443), score: -0.46 ACER2alkaline ceramidase 2 (ENSG00000177076), score: 0.38 ACSL5acyl-CoA synthetase long-chain family member 5 (ENSG00000197142), score: 0.4 ACTR8ARP8 actin-related protein 8 homolog (yeast) (ENSG00000113812), score: 0.54 ADPRHADP-ribosylarginine hydrolase (ENSG00000144843), score: 0.47 AFAP1L2actin filament associated protein 1-like 2 (ENSG00000169129), score: -0.48 AFF4AF4/FMR2 family, member 4 (ENSG00000072364), score: 0.46 AKR1A1aldo-keto reductase family 1, member A1 (aldehyde reductase) (ENSG00000117448), score: 0.42 AMMECR1LAMME chromosomal region gene 1-like (ENSG00000144233), score: 0.61 ANAPC4anaphase promoting complex subunit 4 (ENSG00000053900), score: 0.54 ANGEL2angel homolog 2 (Drosophila) (ENSG00000174606), score: 0.66 ANKRD13Cankyrin repeat domain 13C (ENSG00000118454), score: 0.39 ANKRD16ankyrin repeat domain 16 (ENSG00000134461), score: 0.47 ANO5anoctamin 5 (ENSG00000171714), score: -0.53 AP3B1adaptor-related protein complex 3, beta 1 subunit (ENSG00000132842), score: 0.55 APBA3amyloid beta (A4) precursor protein-binding, family A, member 3 (ENSG00000011132), score: 0.42 ARFGEF1ADP-ribosylation factor guanine nucleotide-exchange factor 1(brefeldin A-inhibited) (ENSG00000066777), score: 0.65 ARFGEF2ADP-ribosylation factor guanine nucleotide-exchange factor 2 (brefeldin A-inhibited) (ENSG00000124198), score: 0.45 ARFIP2ADP-ribosylation factor interacting protein 2 (ENSG00000132254), score: -0.66 ARHGAP12Rho GTPase activating protein 12 (ENSG00000165322), score: 0.41 ARMC7armadillo repeat containing 7 (ENSG00000125449), score: -0.69 ARRDC1arrestin domain containing 1 (ENSG00000197070), score: -0.47 ARSBarylsulfatase B (ENSG00000113273), score: 0.48 ARV1ARV1 homolog (S. cerevisiae) (ENSG00000173409), score: -0.69 ATG4AATG4 autophagy related 4 homolog A (S. cerevisiae) (ENSG00000101844), score: -0.47 ATL2atlastin GTPase 2 (ENSG00000119787), score: 0.39 ATP11AATPase, class VI, type 11A (ENSG00000068650), score: 0.42 ATP6V0A2ATPase, H+ transporting, lysosomal V0 subunit a2 (ENSG00000185344), score: 0.59 ATP9BATPase, class II, type 9B (ENSG00000166377), score: 0.45 ATPAF1ATP synthase mitochondrial F1 complex assembly factor 1 (ENSG00000123472), score: -0.66 ATXN7ataxin 7 (ENSG00000163635), score: 0.41 B3GALT5UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 5 (ENSG00000183778), score: 0.44 B3GALTLbeta 1,3-galactosyltransferase-like (ENSG00000187676), score: -0.46 B4GALT2UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 2 (ENSG00000117411), score: -0.7 BBS2Bardet-Biedl syndrome 2 (ENSG00000125124), score: -0.5 BCL10B-cell CLL/lymphoma 10 (ENSG00000142867), score: 0.59 BPGM2,3-bisphosphoglycerate mutase (ENSG00000172331), score: 0.43 BPNT13'(2'), 5'-bisphosphate nucleotidase 1 (ENSG00000162813), score: 0.53 BRAPBRCA1 associated protein (ENSG00000089234), score: 0.48 C10orf118chromosome 10 open reading frame 118 (ENSG00000165813), score: 0.45 C10orf18chromosome 10 open reading frame 18 (ENSG00000108021), score: 0.45 C10orf46chromosome 10 open reading frame 46 (ENSG00000151893), score: 0.4 C10orf54chromosome 10 open reading frame 54 (ENSG00000107738), score: -0.47 C10orf76chromosome 10 open reading frame 76 (ENSG00000120029), score: -0.48 C11orf74chromosome 11 open reading frame 74 (ENSG00000166352), score: 0.38 C12orf65chromosome 12 open reading frame 65 (ENSG00000130921), score: -0.47 C15orf17chromosome 15 open reading frame 17 (ENSG00000178761), score: -0.56 C17orf28chromosome 17 open reading frame 28 (ENSG00000167861), score: -0.46 C17orf39chromosome 17 open reading frame 39 (ENSG00000141034), score: 0.5 C19orf56chromosome 19 open reading frame 56 (ENSG00000105583), score: -0.52 C1orf128chromosome 1 open reading frame 128 (ENSG00000057757), score: -0.47 C1orf27chromosome 1 open reading frame 27 (ENSG00000157181), score: 0.41 C1orf43chromosome 1 open reading frame 43 (ENSG00000143612), score: -0.57 C1orf58chromosome 1 open reading frame 58 (ENSG00000162819), score: 0.48 C1orf83chromosome 1 open reading frame 83 (ENSG00000116205), score: 0.47 C1orf91chromosome 1 open reading frame 91 (ENSG00000160055), score: 0.66 C21orf63chromosome 21 open reading frame 63 (ENSG00000166979), score: -0.51 C3orf37chromosome 3 open reading frame 37 (ENSG00000183624), score: -0.65 C3orf38chromosome 3 open reading frame 38 (ENSG00000179021), score: 0.39 C3orf75chromosome 3 open reading frame 75 (ENSG00000163832), score: -0.67 C4orf34chromosome 4 open reading frame 34 (ENSG00000163683), score: 0.47 C5orf28chromosome 5 open reading frame 28 (ENSG00000151881), score: 0.54 C5orf41chromosome 5 open reading frame 41 (ENSG00000164463), score: 0.48 C5orf51chromosome 5 open reading frame 51 (ENSG00000205765), score: 0.48 C7orf23chromosome 7 open reading frame 23 (ENSG00000135185), score: 0.51 C9orf82chromosome 9 open reading frame 82 (ENSG00000120159), score: -0.44 CAPRIN2caprin family member 2 (ENSG00000110888), score: -0.52 CASC3cancer susceptibility candidate 3 (ENSG00000108349), score: -0.5 CASP8caspase 8, apoptosis-related cysteine peptidase (ENSG00000064012), score: 0.39 CCDC117coiled-coil domain containing 117 (ENSG00000159873), score: 0.53 CCDC41coiled-coil domain containing 41 (ENSG00000173588), score: 0.44 CCDC51coiled-coil domain containing 51 (ENSG00000164051), score: -0.47 CCDC92coiled-coil domain containing 92 (ENSG00000119242), score: -0.48 CCDC99coiled-coil domain containing 99 (ENSG00000040275), score: -0.44 CCRL1chemokine (C-C motif) receptor-like 1 (ENSG00000129048), score: 0.76 CDK8cyclin-dependent kinase 8 (ENSG00000132964), score: 1 CDKN1Bcyclin-dependent kinase inhibitor 1B (p27, Kip1) (ENSG00000111276), score: -0.44 CETN3centrin, EF-hand protein, 3 (ENSG00000153140), score: 0.47 CGRRF1cell growth regulator with ring finger domain 1 (ENSG00000100532), score: 0.59 CHAC2ChaC, cation transport regulator homolog 2 (E. coli) (ENSG00000143942), score: 0.4 CHMP2Bchromatin modifying protein 2B (ENSG00000083937), score: 0.52 CHUKconserved helix-loop-helix ubiquitous kinase (ENSG00000213341), score: 0.48 CIRBPcold inducible RNA binding protein (ENSG00000099622), score: -0.46 CLEC3BC-type lectin domain family 3, member B (ENSG00000163815), score: -0.55 CNOT6CCR4-NOT transcription complex, subunit 6 (ENSG00000113300), score: 0.48 CNR2cannabinoid receptor 2 (macrophage) (ENSG00000188822), score: 0.7 CNTD1cyclin N-terminal domain containing 1 (ENSG00000176563), score: 0.41 COL10A1collagen, type X, alpha 1 (ENSG00000123500), score: 0.55 COMMD3COMM domain containing 3 (ENSG00000148444), score: 0.43 COPB2coatomer protein complex, subunit beta 2 (beta prime) (ENSG00000184432), score: 0.38 COPEcoatomer protein complex, subunit epsilon (ENSG00000105669), score: -0.52 COQ2coenzyme Q2 homolog, prenyltransferase (yeast) (ENSG00000173085), score: 0.4 CPNE2copine II (ENSG00000140848), score: -0.47 CPSF4Lcleavage and polyadenylation specific factor 4-like (ENSG00000187959), score: 0.48 CPSF6cleavage and polyadenylation specific factor 6, 68kDa (ENSG00000111605), score: -0.58 CROTcarnitine O-octanoyltransferase (ENSG00000005469), score: 0.44 CTNNBIP1catenin, beta interacting protein 1 (ENSG00000178585), score: -0.54 CUL4Acullin 4A (ENSG00000139842), score: 0.41 CUL5cullin 5 (ENSG00000166266), score: 0.42 CYB5R4cytochrome b5 reductase 4 (ENSG00000065615), score: 0.65 CYBRD1cytochrome b reductase 1 (ENSG00000071967), score: -0.73 DAPP1dual adaptor of phosphotyrosine and 3-phosphoinositides (ENSG00000070190), score: 0.48 DARSaspartyl-tRNA synthetase (ENSG00000115866), score: 0.45 DARS2aspartyl-tRNA synthetase 2, mitochondrial (ENSG00000117593), score: 0.54 DCTDdCMP deaminase (ENSG00000129187), score: -0.54 DCUN1D5DCN1, defective in cullin neddylation 1, domain containing 5 (S. cerevisiae) (ENSG00000137692), score: 0.45 DDX41DEAD (Asp-Glu-Ala-Asp) box polypeptide 41 (ENSG00000183258), score: -0.45 DDX47DEAD (Asp-Glu-Ala-Asp) box polypeptide 47 (ENSG00000213782), score: 0.42 DIMT1LDIM1 dimethyladenosine transferase 1-like (S. cerevisiae) (ENSG00000086189), score: -0.67 DNAJC10DnaJ (Hsp40) homolog, subfamily C, member 10 (ENSG00000077232), score: 0.47 DNAJC3DnaJ (Hsp40) homolog, subfamily C, member 3 (ENSG00000102580), score: 0.46 DPH5DPH5 homolog (S. cerevisiae) (ENSG00000117543), score: -0.47 DPM1dolichyl-phosphate mannosyltransferase polypeptide 1, catalytic subunit (ENSG00000000419), score: 0.45 DPP8dipeptidyl-peptidase 8 (ENSG00000074603), score: 0.48 DTNBP1dystrobrevin binding protein 1 (ENSG00000047579), score: 0.45 DUSP14dual specificity phosphatase 14 (ENSG00000161326), score: -0.53 DUSP19dual specificity phosphatase 19 (ENSG00000162999), score: 0.44 DYMdymeclin (ENSG00000141627), score: 0.38 ELK4ELK4, ETS-domain protein (SRF accessory protein 1) (ENSG00000158711), score: 0.47 ELP2elongation protein 2 homolog (S. cerevisiae) (ENSG00000134759), score: 0.42 ENGASEendo-beta-N-acetylglucosaminidase (ENSG00000167280), score: -0.53 ENTPD4ectonucleoside triphosphate diphosphohydrolase 4 (ENSG00000197217), score: 0.7 EPB41erythrocyte membrane protein band 4.1 (elliptocytosis 1, RH-linked) (ENSG00000159023), score: 0.42 ERAL1Era G-protein-like 1 (E. coli) (ENSG00000132591), score: -0.47 EVI5ecotropic viral integration site 5 (ENSG00000067208), score: 0.45 EXOC7exocyst complex component 7 (ENSG00000182473), score: -0.45 FABP2fatty acid binding protein 2, intestinal (ENSG00000145384), score: 0.48 FAM101Afamily with sequence similarity 101, member A (ENSG00000178882), score: -0.44 FAM105Bfamily with sequence similarity 105, member B (ENSG00000154124), score: 0.51 FAM122Bfamily with sequence similarity 122B (ENSG00000156504), score: -0.73 FAM151Bfamily with sequence similarity 151, member B (ENSG00000152380), score: 0.4 FAM160B1family with sequence similarity 160, member B1 (ENSG00000151553), score: 0.53 FAM175Bfamily with sequence similarity 175, member B (ENSG00000165660), score: 0.57 FAM199Xfamily with sequence similarity 199, X-linked (ENSG00000123575), score: -0.64 FAM36Afamily with sequence similarity 36, member A (ENSG00000203667), score: -0.53 FAM76Afamily with sequence similarity 76, member A (ENSG00000009780), score: 0.54 FAM8A1family with sequence similarity 8, member A1 (ENSG00000137414), score: 0.38 FARP1FERM, RhoGEF (ARHGEF) and pleckstrin domain protein 1 (chondrocyte-derived) (ENSG00000152767), score: -0.46 FARS2phenylalanyl-tRNA synthetase 2, mitochondrial (ENSG00000145982), score: 0.49 FBXL15F-box and leucine-rich repeat protein 15 (ENSG00000107872), score: -0.55 FBXL5F-box and leucine-rich repeat protein 5 (ENSG00000118564), score: -0.45 FBXL7F-box and leucine-rich repeat protein 7 (ENSG00000183580), score: -0.52 FBXO3F-box protein 3 (ENSG00000110429), score: 0.44 FBXO30F-box protein 30 (ENSG00000118496), score: 0.46 FBXO7F-box protein 7 (ENSG00000100225), score: -0.51 FBXW4F-box and WD repeat domain containing 4 (ENSG00000107829), score: -0.53 FBXW8F-box and WD repeat domain containing 8 (ENSG00000174989), score: 0.6 FCHO2FCH domain only 2 (ENSG00000157107), score: 0.49 FGD6FYVE, RhoGEF and PH domain containing 6 (ENSG00000180263), score: 0.42 FHL3four and a half LIM domains 3 (ENSG00000183386), score: -0.47 FKTNfukutin (ENSG00000106692), score: 0.43 FMR1fragile X mental retardation 1 (ENSG00000102081), score: -0.46 FRS2fibroblast growth factor receptor substrate 2 (ENSG00000166225), score: 0.53 GABARAPL2GABA(A) receptor-associated protein-like 2 (ENSG00000034713), score: -0.58 GADD45Bgrowth arrest and DNA-damage-inducible, beta (ENSG00000099860), score: -0.54 GALNT1UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 1 (GalNAc-T1) (ENSG00000141429), score: 0.42 GAPVD1GTPase activating protein and VPS9 domains 1 (ENSG00000165219), score: 0.46 GBA2glucosidase, beta (bile acid) 2 (ENSG00000070610), score: -0.52 GCLMglutamate-cysteine ligase, modifier subunit (ENSG00000023909), score: 0.4 GDE1glycerophosphodiester phosphodiesterase 1 (ENSG00000006007), score: 0.51 GGPS1geranylgeranyl diphosphate synthase 1 (ENSG00000152904), score: -0.56 GIT2G protein-coupled receptor kinase interacting ArfGAP 2 (ENSG00000139436), score: -0.49 GLIPR2GLI pathogenesis-related 2 (ENSG00000122694), score: -0.45 GM2AGM2 ganglioside activator (ENSG00000196743), score: 0.47 GNA12guanine nucleotide binding protein (G protein) alpha 12 (ENSG00000146535), score: 0.47 GNA13guanine nucleotide binding protein (G protein), alpha 13 (ENSG00000120063), score: 0.5 GOLGA4golgin A4 (ENSG00000144674), score: 0.49 GPATCH2G patch domain containing 2 (ENSG00000092978), score: 0.39 GPR180G protein-coupled receptor 180 (ENSG00000152749), score: 0.76 GRPEL2GrpE-like 2, mitochondrial (E. coli) (ENSG00000164284), score: 0.4 GXYLT1glucoside xylosyltransferase 1 (ENSG00000151233), score: 0.62 H1FXH1 histone family, member X (ENSG00000184897), score: -0.88 HAPLN3hyaluronan and proteoglycan link protein 3 (ENSG00000140511), score: -0.46 HBP1HMG-box transcription factor 1 (ENSG00000105856), score: 0.4 HDAC8histone deacetylase 8 (ENSG00000147099), score: -0.53 HDDC2HD domain containing 2 (ENSG00000111906), score: -0.69 HDHD3haloacid dehalogenase-like hydrolase domain containing 3 (ENSG00000119431), score: -0.5 HELBhelicase (DNA) B (ENSG00000127311), score: 0.48 HINFPhistone H4 transcription factor (ENSG00000172273), score: -0.72 HIPK3homeodomain interacting protein kinase 3 (ENSG00000110422), score: 0.58 HIVEP1human immunodeficiency virus type I enhancer binding protein 1 (ENSG00000095951), score: 0.39 HMBOX1homeobox containing 1 (ENSG00000147421), score: 0.52 HNRNPRheterogeneous nuclear ribonucleoprotein R (ENSG00000125944), score: -0.64 HSF1heat shock transcription factor 1 (ENSG00000185122), score: -0.47 IDEinsulin-degrading enzyme (ENSG00000119912), score: 0.41 IFFO1intermediate filament family orphan 1 (ENSG00000010295), score: -0.58 IGHMBP2immunoglobulin mu binding protein 2 (ENSG00000132740), score: -0.61 IL12RB2interleukin 12 receptor, beta 2 (ENSG00000081985), score: 0.42 ING1inhibitor of growth family, member 1 (ENSG00000153487), score: 0.45 INO80DINO80 complex subunit D (ENSG00000114933), score: 0.39 INPP5Binositol polyphosphate-5-phosphatase, 75kDa (ENSG00000204084), score: 0.39 INTS7integrator complex subunit 7 (ENSG00000143493), score: 0.39 ITM2Aintegral membrane protein 2A (ENSG00000078596), score: -0.53 ITM2Bintegral membrane protein 2B (ENSG00000136156), score: 0.4 IVNS1ABPinfluenza virus NS1A binding protein (ENSG00000116679), score: 0.43 IWS1IWS1 homolog (S. cerevisiae) (ENSG00000163166), score: -0.45 JAK2Janus kinase 2 (ENSG00000096968), score: 0.47 KCNMB1potassium large conductance calcium-activated channel, subfamily M, beta member 1 (ENSG00000145936), score: -0.45 KCNRGpotassium channel regulator (ENSG00000198553), score: -0.52 KIAA0020KIAA0020 (ENSG00000080608), score: 0.41 KIAA0100KIAA0100 (ENSG00000007202), score: 0.58 KIAA0174KIAA0174 (ENSG00000182149), score: -0.45 KIAA2018KIAA2018 (ENSG00000176542), score: 0.54 KLF8Kruppel-like factor 8 (ENSG00000102349), score: -0.51 KRT80keratin 80 (ENSG00000167767), score: 0.46 L3MBTL2l(3)mbt-like 2 (Drosophila) (ENSG00000100395), score: -0.45 LACE1lactation elevated 1 (ENSG00000135537), score: 0.44 LARP4BLa ribonucleoprotein domain family, member 4B (ENSG00000107929), score: 0.42 LATS1LATS, large tumor suppressor, homolog 1 (Drosophila) (ENSG00000131023), score: 0.44 LCLAT1lysocardiolipin acyltransferase 1 (ENSG00000172954), score: 0.59 LIFRleukemia inhibitory factor receptor alpha (ENSG00000113594), score: 0.47 LMBRD2LMBR1 domain containing 2 (ENSG00000164187), score: 0.42 LOC100131693eukaryotic translation initiation factor 4E pseudogene (ENSG00000151247), score: 0.57 LOC100294349similar to Parkinson disease 7 domain containing 1 (ENSG00000177225), score: -0.52 LPGAT1lysophosphatidylglycerol acyltransferase 1 (ENSG00000123684), score: 0.6 LPIN1lipin 1 (ENSG00000134324), score: 0.42 LRSAM1leucine rich repeat and sterile alpha motif containing 1 (ENSG00000148356), score: -0.55 LRTM1leucine-rich repeats and transmembrane domains 1 (ENSG00000144771), score: 0.5 LSM10LSM10, U7 small nuclear RNA associated (ENSG00000181817), score: -0.45 MAP1Dmethionine aminopeptidase 1D (ENSG00000172878), score: 0.63 MAP3K1mitogen-activated protein kinase kinase kinase 1 (ENSG00000095015), score: 0.42 MAP3K14mitogen-activated protein kinase kinase kinase 14 (ENSG00000006062), score: -0.55 MARCH5membrane-associated ring finger (C3HC4) 5 (ENSG00000198060), score: 0.69 MARCH6membrane-associated ring finger (C3HC4) 6 (ENSG00000145495), score: 0.48 MBIPMAP3K12 binding inhibitory protein 1 (ENSG00000151332), score: -0.56 MCM3APminichromosome maintenance complex component 3 associated protein (ENSG00000160294), score: -0.68 MCRS1microspherule protein 1 (ENSG00000187778), score: -0.51 MDM4Mdm4 p53 binding protein homolog (mouse) (ENSG00000198625), score: 0.51 MEAF6MYST/Esa1-associated factor 6 (ENSG00000163875), score: -0.66 MED1mediator complex subunit 1 (ENSG00000125686), score: 0.42 MED28mediator complex subunit 28 (ENSG00000118579), score: 0.8 MEP1Ameprin A, alpha (PABA peptide hydrolase) (ENSG00000112818), score: 0.41 MEP1Bmeprin A, beta (ENSG00000141434), score: 0.41 METT10Dmethyltransferase 10 domain containing (ENSG00000127804), score: -0.56 METTL6methyltransferase like 6 (ENSG00000206562), score: 0.39 MFN1mitofusin 1 (ENSG00000171109), score: 0.57 MGST2microsomal glutathione S-transferase 2 (ENSG00000085871), score: -0.48 MIA3melanoma inhibitory activity family, member 3 (ENSG00000154305), score: 0.58 MICAL1microtubule associated monoxygenase, calponin and LIM domain containing 1 (ENSG00000135596), score: -0.56 MICALL2MICAL-like 2 (ENSG00000164877), score: -0.54 MINPP1multiple inositol-polyphosphate phosphatase 1 (ENSG00000107789), score: 0.51 MIPOL1mirror-image polydactyly 1 (ENSG00000151338), score: 0.63 MKS1Meckel syndrome, type 1 (ENSG00000011143), score: -0.52 MLXMAX-like protein X (ENSG00000108788), score: -0.45 MMAAmethylmalonic aciduria (cobalamin deficiency) cblA type (ENSG00000151611), score: 0.4 MMP13matrix metallopeptidase 13 (collagenase 3) (ENSG00000137745), score: 0.39 MPP6membrane protein, palmitoylated 6 (MAGUK p55 subfamily member 6) (ENSG00000105926), score: 0.47 MRASmuscle RAS oncogene homolog (ENSG00000158186), score: -0.45 MRPL21mitochondrial ribosomal protein L21 (ENSG00000197345), score: -0.57 MRPS11mitochondrial ribosomal protein S11 (ENSG00000181991), score: -0.56 MRPS25mitochondrial ribosomal protein S25 (ENSG00000131368), score: -0.45 MTFMTmitochondrial methionyl-tRNA formyltransferase (ENSG00000103707), score: -0.46 MTMR10myotubularin related protein 10 (ENSG00000166912), score: -0.45 MTMR6myotubularin related protein 6 (ENSG00000139505), score: 0.55 MUTYHmutY homolog (E. coli) (ENSG00000132781), score: -0.55 NAA25N(alpha)-acetyltransferase 25, NatB auxiliary subunit (ENSG00000111300), score: 0.39 NAA35N(alpha)-acetyltransferase 35, NatC auxiliary subunit (ENSG00000135040), score: 0.57 NCBP1nuclear cap binding protein subunit 1, 80kDa (ENSG00000136937), score: 0.57 ND3NADH dehydrogenase, subunit 3 (complex I) (ENSG00000198840), score: -0.62 NDUFA6NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 6, 14kDa (ENSG00000184983), score: 0.4 NECAB3N-terminal EF-hand calcium binding protein 3 (ENSG00000125967), score: -0.53 NEDD4neural precursor cell expressed, developmentally down-regulated 4 (ENSG00000069869), score: 0.91 NEIL1nei endonuclease VIII-like 1 (E. coli) (ENSG00000140398), score: -0.5 NFAT5nuclear factor of activated T-cells 5, tonicity-responsive (ENSG00000102908), score: 0.42 NFRKBnuclear factor related to kappaB binding protein (ENSG00000170322), score: -0.51 NHEDC2Na+/H+ exchanger domain containing 2 (ENSG00000164038), score: -0.49 NHLRC2NHL repeat containing 2 (ENSG00000196865), score: 0.39 NINJ2ninjurin 2 (ENSG00000171840), score: -0.47 NSL1NSL1, MIND kinetochore complex component, homolog (S. cerevisiae) (ENSG00000117697), score: -0.88 NSMAFneutral sphingomyelinase (N-SMase) activation associated factor (ENSG00000035681), score: 0.46 NSUN2NOP2/Sun domain family, member 2 (ENSG00000037474), score: 0.54 NTHL1nth endonuclease III-like 1 (E. coli) (ENSG00000065057), score: -0.61 NUDCD1NudC domain containing 1 (ENSG00000120526), score: 0.43 NUDT19nudix (nucleoside diphosphate linked moiety X)-type motif 19 (ENSG00000213965), score: 0.59 NUDT4nudix (nucleoside diphosphate linked moiety X)-type motif 4 (ENSG00000173598), score: 0.54 OBFC2Aoligonucleotide/oligosaccharide-binding fold containing 2A (ENSG00000173559), score: 0.45 ORC4Lorigin recognition complex, subunit 4-like (S. cerevisiae) (ENSG00000115947), score: -0.47 OSGIN2oxidative stress induced growth inhibitor family member 2 (ENSG00000164823), score: -0.57 OXNAD1oxidoreductase NAD-binding domain containing 1 (ENSG00000154814), score: 0.51 P4HTMprolyl 4-hydroxylase, transmembrane (endoplasmic reticulum) (ENSG00000178467), score: -0.51 PAK1IP1PAK1 interacting protein 1 (ENSG00000111845), score: 0.65 PALB2partner and localizer of BRCA2 (ENSG00000083093), score: -0.63 PAPD5PAP associated domain containing 5 (ENSG00000121274), score: 0.41 PAQR9progestin and adipoQ receptor family member IX (ENSG00000188582), score: 0.48 PAXIP1PAX interacting (with transcription-activation domain) protein 1 (ENSG00000157212), score: 0.41 PCBD2pterin-4 alpha-carbinolamine dehydratase/dimerization cofactor of hepatocyte nuclear factor 1 alpha (TCF1) 2 (ENSG00000132570), score: 0.39 PDCD5programmed cell death 5 (ENSG00000105185), score: 0.39 PELI2pellino homolog 2 (Drosophila) (ENSG00000139946), score: 0.42 PEX10peroxisomal biogenesis factor 10 (ENSG00000157911), score: -0.68 PEX11Aperoxisomal biogenesis factor 11 alpha (ENSG00000166821), score: 0.46 PEX13peroxisomal biogenesis factor 13 (ENSG00000162928), score: 0.43 PFDN1prefoldin subunit 1 (ENSG00000113068), score: 0.41 PGAP3post-GPI attachment to proteins 3 (ENSG00000161395), score: -0.61 PHKA2phosphorylase kinase, alpha 2 (liver) (ENSG00000044446), score: -0.51 PHTF2putative homeodomain transcription factor 2 (ENSG00000006576), score: 0.41 PIGPphosphatidylinositol glycan anchor biosynthesis, class P (ENSG00000185808), score: 0.39 PIP4K2Aphosphatidylinositol-5-phosphate 4-kinase, type II, alpha (ENSG00000150867), score: -0.47 PKD2L2polycystic kidney disease 2-like 2 (ENSG00000078795), score: 0.47 PLEKHN1pleckstrin homology domain containing, family N member 1 (ENSG00000187583), score: 0.79 PMS1PMS1 postmeiotic segregation increased 1 (S. cerevisiae) (ENSG00000064933), score: -0.44 POLG2polymerase (DNA directed), gamma 2, accessory subunit (ENSG00000136480), score: -0.47 POLR1Cpolymerase (RNA) I polypeptide C, 30kDa (ENSG00000171453), score: 0.46 POLR2Cpolymerase (RNA) II (DNA directed) polypeptide C, 33kDa (ENSG00000102978), score: -0.45 POLR3Gpolymerase (RNA) III (DNA directed) polypeptide G (32kD) (ENSG00000113356), score: 0.52 POLR3Kpolymerase (RNA) III (DNA directed) polypeptide K, 12.3 kDa (ENSG00000161980), score: -0.6 PPA2pyrophosphatase (inorganic) 2 (ENSG00000138777), score: 0.38 PPM1Bprotein phosphatase, Mg2+/Mn2+ dependent, 1B (ENSG00000138032), score: 0.62 PPP1R7protein phosphatase 1, regulatory (inhibitor) subunit 7 (ENSG00000115685), score: -0.61 PPP2R5Aprotein phosphatase 2, regulatory subunit B', alpha (ENSG00000066027), score: 0.47 PPP6R2protein phosphatase 6, regulatory subunit 2 (ENSG00000100239), score: -0.56 PQLC1PQ loop repeat containing 1 (ENSG00000122490), score: -0.54 PQLC3PQ loop repeat containing 3 (ENSG00000162976), score: -0.56 PRKD3protein kinase D3 (ENSG00000115825), score: 0.43 PROX2prospero homeobox 2 (ENSG00000119608), score: 0.49 PRRG1proline rich Gla (G-carboxyglutamic acid) 1 (ENSG00000130962), score: -0.56 PSEN1presenilin 1 (ENSG00000080815), score: 0.39 PSMA4proteasome (prosome, macropain) subunit, alpha type, 4 (ENSG00000041357), score: 0.44 PSMC2proteasome (prosome, macropain) 26S subunit, ATPase, 2 (ENSG00000161057), score: 0.44 PSMD12proteasome (prosome, macropain) 26S subunit, non-ATPase, 12 (ENSG00000197170), score: 0.63 PSME4proteasome (prosome, macropain) activator subunit 4 (ENSG00000068878), score: 0.4 PSMG3proteasome (prosome, macropain) assembly chaperone 3 (ENSG00000157778), score: -0.63 PSTPIP2proline-serine-threonine phosphatase interacting protein 2 (ENSG00000152229), score: 0.59 PTGR2prostaglandin reductase 2 (ENSG00000140043), score: 0.43 PTPLAD2protein tyrosine phosphatase-like A domain containing 2 (ENSG00000188921), score: 0.51 PUS10pseudouridylate synthase 10 (ENSG00000162927), score: 0.46 RAB22ARAB22A, member RAS oncogene family (ENSG00000124209), score: 0.58 RAB3IPRAB3A interacting protein (rabin3) (ENSG00000127328), score: 0.39 RAD51RAD51 homolog (RecA homolog, E. coli) (S. cerevisiae) (ENSG00000051180), score: 0.43 RALGAPA2Ral GTPase activating protein, alpha subunit 2 (catalytic) (ENSG00000188559), score: 0.44 RASD1RAS, dexamethasone-induced 1 (ENSG00000108551), score: -0.47 RBM14RNA binding motif protein 14 (ENSG00000239306), score: -0.48 RBM45RNA binding motif protein 45 (ENSG00000155636), score: 0.4 RC3H1ring finger and CCCH-type zinc finger domains 1 (ENSG00000135870), score: 0.39 RDM1RAD52 motif 1 (ENSG00000187456), score: 0.42 RDXradixin (ENSG00000137710), score: 0.53 RFESDRieske (Fe-S) domain containing (ENSG00000175449), score: 0.68 RFWD3ring finger and WD repeat domain 3 (ENSG00000168411), score: 0.52 RNF139ring finger protein 139 (ENSG00000170881), score: 0.72 RP2retinitis pigmentosa 2 (X-linked recessive) (ENSG00000102218), score: 0.43 RPRD2regulation of nuclear pre-mRNA domain containing 2 (ENSG00000163125), score: -0.47 RPS19BP1ribosomal protein S19 binding protein 1 (ENSG00000187051), score: -0.62 RPUSD2RNA pseudouridylate synthase domain containing 2 (ENSG00000166133), score: -0.57 RRAGCRas-related GTP binding C (ENSG00000116954), score: 0.55 SAP30BPSAP30 binding protein (ENSG00000161526), score: -0.67 SART3squamous cell carcinoma antigen recognized by T cells 3 (ENSG00000075856), score: -0.57 SCLYselenocysteine lyase (ENSG00000132330), score: 0.38 SEC23IPSEC23 interacting protein (ENSG00000107651), score: 0.55 SEC31BSEC31 homolog B (S. cerevisiae) (ENSG00000075826), score: -0.65 SENP6SUMO1/sentrin specific peptidase 6 (ENSG00000112701), score: 0.42 SEP1515 kDa selenoprotein (ENSG00000183291), score: 0.51 SEPSECSSep (O-phosphoserine) tRNA:Sec (selenocysteine) tRNA synthase (ENSG00000109618), score: 0.38 SEPT6septin 6 (ENSG00000125354), score: -0.45 SERINC3serine incorporator 3 (ENSG00000132824), score: 0.55 SFMBT2Scm-like with four mbt domains 2 (ENSG00000198879), score: -0.5 SGPP1sphingosine-1-phosphate phosphatase 1 (ENSG00000126821), score: 0.39 SGSM3small G protein signaling modulator 3 (ENSG00000100359), score: -0.57 SIRT3sirtuin 3 (ENSG00000142082), score: 0.41 SLC17A8solute carrier family 17 (sodium-dependent inorganic phosphate cotransporter), member 8 (ENSG00000179520), score: 0.41 SLC18A1solute carrier family 18 (vesicular monoamine), member 1 (ENSG00000036565), score: 0.4 SLC25A16solute carrier family 25 (mitochondrial carrier; Graves disease autoantigen), member 16 (ENSG00000122912), score: 0.44 SLC25A28solute carrier family 25, member 28 (ENSG00000155287), score: -0.66 SLC25A29solute carrier family 25, member 29 (ENSG00000197119), score: -0.55 SLC25A40solute carrier family 25, member 40 (ENSG00000075303), score: -0.46 SLC25A46solute carrier family 25, member 46 (ENSG00000164209), score: 0.38 SLC30A6solute carrier family 30 (zinc transporter), member 6 (ENSG00000152683), score: 0.49 SLC6A6solute carrier family 6 (neurotransmitter transporter, taurine), member 6 (ENSG00000131389), score: 0.48 SLC9A8solute carrier family 9 (sodium/hydrogen exchanger), member 8 (ENSG00000197818), score: 0.45 SMAD5SMAD family member 5 (ENSG00000113658), score: 0.39 SMARCA1SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 1 (ENSG00000102038), score: -0.74 SMC5structural maintenance of chromosomes 5 (ENSG00000198887), score: 0.45 SMNDC1survival motor neuron domain containing 1 (ENSG00000119953), score: 0.49 SNX11sorting nexin 11 (ENSG00000002919), score: -0.56 SNX17sorting nexin 17 (ENSG00000115234), score: -0.58 SPATS2Lspermatogenesis associated, serine-rich 2-like (ENSG00000196141), score: -0.46 SSBP1single-stranded DNA binding protein 1 (ENSG00000106028), score: -0.55 SSPNsarcospan (Kras oncogene-associated gene) (ENSG00000123096), score: 0.85 ST6GALNAC4ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 4 (ENSG00000136840), score: -0.51 ST7suppression of tumorigenicity 7 (ENSG00000004866), score: -0.47 STK17Bserine/threonine kinase 17b (ENSG00000081320), score: 0.43 TADA3transcriptional adaptor 3 (ENSG00000171148), score: -0.46 TAF1ATATA box binding protein (TBP)-associated factor, RNA polymerase I, A, 48kDa (ENSG00000143498), score: 0.59 TAF1BTATA box binding protein (TBP)-associated factor, RNA polymerase I, B, 63kDa (ENSG00000115750), score: 0.39 TBCELtubulin folding cofactor E-like (ENSG00000154114), score: 0.59 TCP11L1t-complex 11 (mouse)-like 1 (ENSG00000176148), score: -0.64 TCP11L2t-complex 11 (mouse)-like 2 (ENSG00000166046), score: 0.41 TEAD4TEA domain family member 4 (ENSG00000197905), score: -0.47 TECtec protein tyrosine kinase (ENSG00000135605), score: 0.38 TELO2TEL2, telomere maintenance 2, homolog (S. cerevisiae) (ENSG00000100726), score: -0.62 TGFBRAP1transforming growth factor, beta receptor associated protein 1 (ENSG00000135966), score: 0.44 TJAP1tight junction associated protein 1 (peripheral) (ENSG00000137221), score: -0.47 TM2D2TM2 domain containing 2 (ENSG00000169490), score: 0.48 TM9SF2transmembrane 9 superfamily member 2 (ENSG00000125304), score: 0.4 TMBIM4transmembrane BAX inhibitor motif containing 4 (ENSG00000155957), score: 0.52 TMCC1transmembrane and coiled-coil domain family 1 (ENSG00000172765), score: 0.43 TMCC3transmembrane and coiled-coil domain family 3 (ENSG00000057704), score: 0.39 TMEM106Btransmembrane protein 106B (ENSG00000106460), score: 0.56 TMEM135transmembrane protein 135 (ENSG00000166575), score: 0.5 TMEM14Atransmembrane protein 14A (ENSG00000096092), score: -0.7 TMEM64transmembrane protein 64 (ENSG00000180694), score: 0.74 TMF1TATA element modulatory factor 1 (ENSG00000144747), score: 0.43 TMX1thioredoxin-related transmembrane protein 1 (ENSG00000139921), score: 0.54 TNFAIP8tumor necrosis factor, alpha-induced protein 8 (ENSG00000145779), score: 0.42 TNFAIP8L3tumor necrosis factor, alpha-induced protein 8-like 3 (ENSG00000183578), score: -0.47 TOMM70Atranslocase of outer mitochondrial membrane 70 homolog A (S. cerevisiae) (ENSG00000154174), score: 0.53 TOP3Atopoisomerase (DNA) III alpha (ENSG00000177302), score: -0.45 TOPORStopoisomerase I binding, arginine/serine-rich (ENSG00000197579), score: 0.48 TSPAN3tetraspanin 3 (ENSG00000140391), score: -0.44 TTC39Ctetratricopeptide repeat domain 39C (ENSG00000168234), score: 0.42 TUBE1tubulin, epsilon 1 (ENSG00000074935), score: -0.59 TULP4tubby like protein 4 (ENSG00000130338), score: 0.44 TXNDC16thioredoxin domain containing 16 (ENSG00000087301), score: 0.54 TXNDC9thioredoxin domain containing 9 (ENSG00000115514), score: 0.71 TYW3tRNA-yW synthesizing protein 3 homolog (S. cerevisiae) (ENSG00000162623), score: -0.62 UBE2Aubiquitin-conjugating enzyme E2A (RAD6 homolog) (ENSG00000077721), score: -0.46 UBE2Wubiquitin-conjugating enzyme E2W (putative) (ENSG00000104343), score: 0.54 UBLCP1ubiquitin-like domain containing CTD phosphatase 1 (ENSG00000164332), score: 0.54 UBR2ubiquitin protein ligase E3 component n-recognin 2 (ENSG00000024048), score: 0.62 UBXN2AUBX domain protein 2A (ENSG00000173960), score: 0.48 UHRF1BP1LUHRF1 binding protein 1-like (ENSG00000111647), score: 0.46 URB2URB2 ribosome biogenesis 2 homolog (S. cerevisiae) (ENSG00000135763), score: 0.44 USP47ubiquitin specific peptidase 47 (ENSG00000170242), score: 0.55 USP8ubiquitin specific peptidase 8 (ENSG00000138592), score: 0.45 USPL1ubiquitin specific peptidase like 1 (ENSG00000132952), score: 0.48 VPS13Avacuolar protein sorting 13 homolog A (S. cerevisiae) (ENSG00000197969), score: 0.49 VPS26Avacuolar protein sorting 26 homolog A (S. pombe) (ENSG00000122958), score: 0.56 VPS45vacuolar protein sorting 45 homolog (S. cerevisiae) (ENSG00000136631), score: -0.54 VPS54vacuolar protein sorting 54 homolog (S. cerevisiae) (ENSG00000143952), score: 0.39 WBP1WW domain binding protein 1 (ENSG00000115274), score: -0.53 WBP4WW domain binding protein 4 (formin binding protein 21) (ENSG00000120688), score: 0.55 WDR36WD repeat domain 36 (ENSG00000134987), score: 0.65 WDR59WD repeat domain 59 (ENSG00000103091), score: -0.47 WDR89WD repeat domain 89 (ENSG00000140006), score: 0.9 WHSC2Wolf-Hirschhorn syndrome candidate 2 (ENSG00000185049), score: -0.47 WRBtryptophan rich basic protein (ENSG00000182093), score: -0.52 XRCC4X-ray repair complementing defective repair in Chinese hamster cells 4 (ENSG00000152422), score: 0.52 YAF2YY1 associated factor 2 (ENSG00000015153), score: 0.4 YTHDF2YTH domain family, member 2 (ENSG00000198492), score: -0.53 ZBTB41zinc finger and BTB domain containing 41 (ENSG00000177888), score: 0.62 ZC3H12Czinc finger CCCH-type containing 12C (ENSG00000149289), score: 0.45 ZC3H7Azinc finger CCCH-type containing 7A (ENSG00000122299), score: 0.94 ZC4H2zinc finger, C4H2 domain containing (ENSG00000126970), score: -0.49 ZCCHC4zinc finger, CCHC domain containing 4 (ENSG00000168228), score: -0.48 ZCCHC8zinc finger, CCHC domain containing 8 (ENSG00000033030), score: 0.48 ZMYND19zinc finger, MYND-type containing 19 (ENSG00000165724), score: -0.53 ZNF518Bzinc finger protein 518B (ENSG00000178163), score: -0.48 ZNF750zinc finger protein 750 (ENSG00000141579), score: 0.57
| Id | species | tissue | sex | individual |
|---|---|---|---|---|
| hsa_br_m7_ca1 | hsa | br | m | 7 |
| ptr_br_m1_ca1 | ptr | br | m | 1 |
| hsa_br_m3_ca1 | hsa | br | m | 3 |
| ptr_br_m2_ca1 | ptr | br | m | 2 |
| hsa_kd_m1_ca1 | hsa | kd | m | 1 |
| ppa_br_f1_ca1 | ppa | br | f | 1 |
| ptr_br_m3_ca1 | ptr | br | m | 3 |
| ggo_br_m_ca1 | ggo | br | m | _ |
| mmu_br_m1_ca1 | mmu | br | m | 1 |
| mmu_br_m2_ca1 | mmu | br | m | 2 |
| mmu_cb_m1_ca1 | mmu | cb | m | 1 |
| mmu_br_f_ca1 | mmu | br | f | _ |
| mmu_cb_f_ca1 | mmu | cb | f | _ |
| mmu_cb_m2_ca1 | mmu | cb | m | 2 |
| mmu_ht_f_ca1 | mmu | ht | f | _ |
| mmu_ts_m1_ca1 | mmu | ts | m | 1 |
| mmu_ht_m1_ca1 | mmu | ht | m | 1 |
| mmu_ht_m2_ca1 | mmu | ht | m | 2 |
| mmu_ts_m2_ca1 | mmu | ts | m | 2 |
| mmu_kd_f_ca1 | mmu | kd | f | _ |
| mmu_kd_m2_ca1 | mmu | kd | m | 2 |
| mmu_kd_m1_ca1 | mmu | kd | m | 1 |
| mmu_lv_m1_ca1 | mmu | lv | m | 1 |
| mmu_lv_f_ca1 | mmu | lv | f | _ |
| mmu_lv_m2_ca1 | mmu | lv | m | 2 |