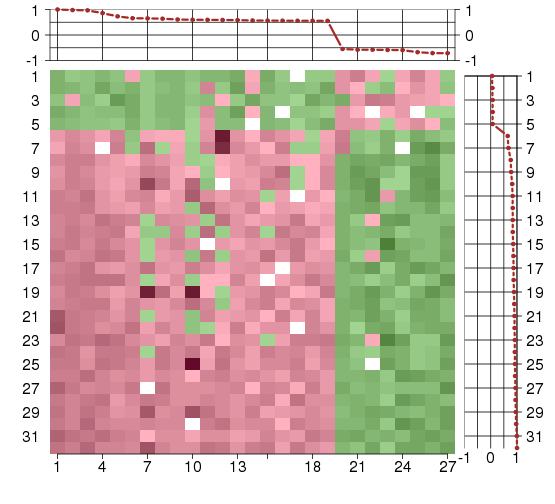

Under-expression is coded with green,
over-expression with red color.



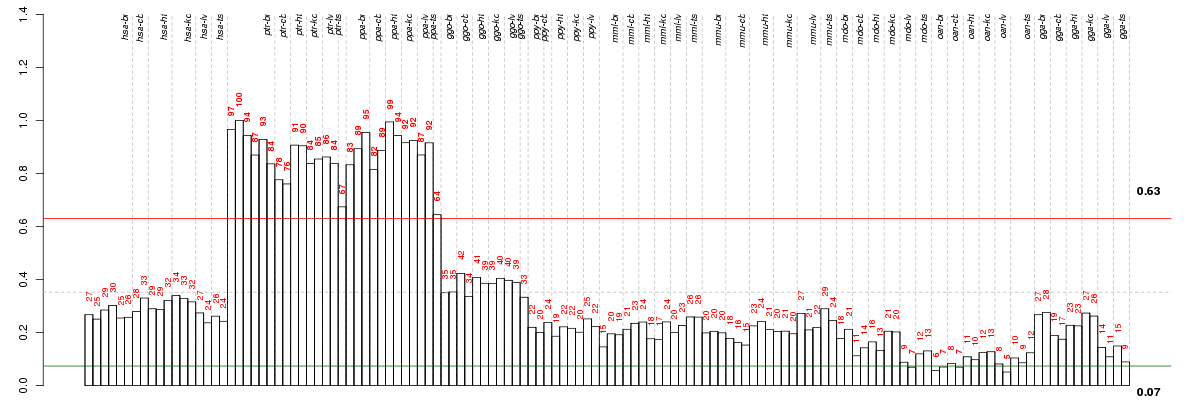
BEST1bestrophin 1 (ENSG00000167995), score: 0.61 C16orf73chromosome 16 open reading frame 73 (ENSG00000162039), score: 0.97 CELcarboxyl ester lipase (bile salt-stimulated lipase) (ENSG00000170835), score: 0.64 CNTD1cyclin N-terminal domain containing 1 (ENSG00000176563), score: -0.55 COL7A1collagen, type VII, alpha 1 (ENSG00000114270), score: 0.57 DGUOKdeoxyguanosine kinase (ENSG00000114956), score: 0.57 DRG2developmentally regulated GTP binding protein 2 (ENSG00000108591), score: 0.56 ENTPD1ectonucleoside triphosphate diphosphohydrolase 1 (ENSG00000138185), score: 1 GRTP1growth hormone regulated TBC protein 1 (ENSG00000139835), score: 0.73 GTF3Ageneral transcription factor IIIA (ENSG00000122034), score: 0.56 HEMK1HemK methyltransferase family member 1 (ENSG00000114735), score: -0.58 HNRNPUheterogeneous nuclear ribonucleoprotein U (scaffold attachment factor A) (ENSG00000153187), score: -0.59 HSCBHscB iron-sulfur cluster co-chaperone homolog (E. coli) (ENSG00000100209), score: -0.67 IMPG1interphotoreceptor matrix proteoglycan 1 (ENSG00000112706), score: 0.6 KANK3KN motif and ankyrin repeat domains 3 (ENSG00000186994), score: 0.86 MAP3K15mitogen-activated protein kinase kinase kinase 15 (ENSG00000180815), score: 0.66 MKKSMcKusick-Kaufman syndrome (ENSG00000125863), score: 0.58 MRP63mitochondrial ribosomal protein 63 (ENSG00000173141), score: -0.71 MRPS25mitochondrial ribosomal protein S25 (ENSG00000131368), score: -0.58 POLG2polymerase (DNA directed), gamma 2, accessory subunit (ENSG00000136480), score: 0.98 POLR3Bpolymerase (RNA) III (DNA directed) polypeptide B (ENSG00000013503), score: -0.59 PRDM15PR domain containing 15 (ENSG00000141956), score: -0.71 RBM6RNA binding motif protein 6 (ENSG00000004534), score: 0.55 SFPQsplicing factor proline/glutamine-rich (ENSG00000116560), score: 0.56 SYCP2Lsynaptonemal complex protein 2-like (ENSG00000153157), score: 0.65 TNFAIP8L3tumor necrosis factor, alpha-induced protein 8-like 3 (ENSG00000183578), score: 0.59 ZBTB25zinc finger and BTB domain containing 25 (ENSG00000089775), score: 0.59
| Id | species | tissue | sex | individual |
|---|---|---|---|---|
| oan_lv_f_ca1 | oan | lv | f | _ |
| oan_br_m_ca1 | oan | br | m | _ |
| mdo_lv_f_ca1 | mdo | lv | f | _ |
| oan_cb_f_ca1 | oan | cb | f | _ |
| oan_br_f_ca1 | oan | br | f | _ |
| ppa_ts_m_ca1 | ppa | ts | m | _ |
| ptr_ts_m_ca1 | ptr | ts | m | _ |
| ptr_cb_f_ca1 | ptr | cb | f | _ |
| ptr_cb_m_ca1 | ptr | cb | m | _ |
| ppa_cb_m_ca1 | ppa | cb | m | _ |
| ppa_br_m_ca1 | ppa | br | m | _ |
| ptr_br_f_ca1 | ptr | br | f | _ |
| ptr_kd_m_ca1 | ptr | kd | m | _ |
| ptr_lv_f_ca1 | ptr | lv | f | _ |
| ptr_kd_f_ca1 | ptr | kd | f | _ |
| ptr_lv_m_ca1 | ptr | lv | m | _ |
| ptr_br_m4_ca1 | ptr | br | m | 4 |
| ppa_lv_m_ca1 | ppa | lv | m | _ |
| ppa_cb_f_ca1 | ppa | cb | f | _ |
| ppa_br_f1_ca1 | ppa | br | f | 1 |
| ptr_ht_f_ca1 | ptr | ht | f | _ |
| ptr_ht_m_ca1 | ptr | ht | m | _ |
| ppa_lv_f_ca1 | ppa | lv | f | _ |
| ppa_kd_m_ca1 | ppa | kd | m | _ |
| ppa_kd_f_ca1 | ppa | kd | f | _ |
| ptr_br_m5_ca1 | ptr | br | m | 5 |
| ppa_ht_f_ca1 | ppa | ht | f | _ |
| ptr_br_m1_ca1 | ptr | br | m | 1 |
| ppa_br_f2_ca1 | ppa | br | f | 2 |
| ptr_br_m3_ca1 | ptr | br | m | 3 |
| ppa_ht_m_ca1 | ppa | ht | m | _ |
| ptr_br_m2_ca1 | ptr | br | m | 2 |