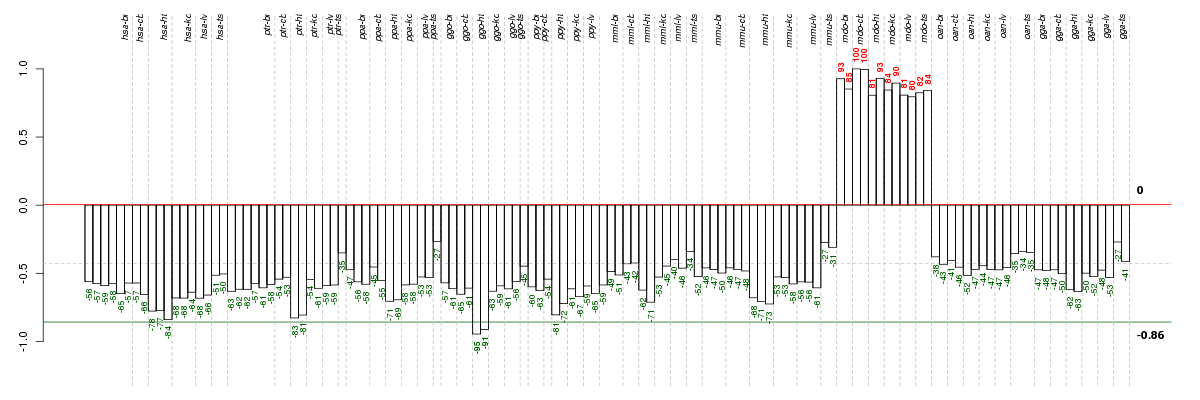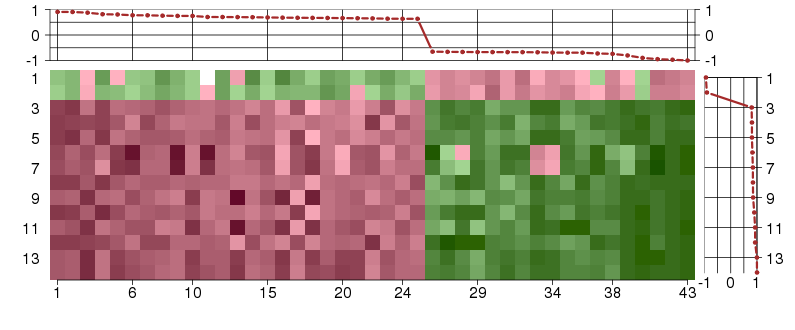

Under-expression is coded with green,
over-expression with red color.


membrane
Double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
cell
The basic structural and functional unit of all organisms. Includes the plasma membrane and any external encapsulating structures such as the cell wall and cell envelope.
membrane part
Any constituent part of a membrane, a double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
respiratory chain
The protein complexes that form the electron transport system (the respiratory chain), associated with a cell membrane, usually the plasma membrane (in prokaryotes) or the inner mitochondrial membrane (on eukaryotes). The respiratory chain complexes transfer electrons from an electron donor to an electron acceptor and are associated with a proton pump to create a transmembrane electrochemical gradient.
all
NA
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
membrane part
Any constituent part of a membrane, a double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.

| Id | Pvalue | ExpCount | Count | Size | Term |
|---|---|---|---|---|---|
| 05012 | 2.826e-04 | 0.2443 | 5 | 27 | Parkinson's disease |
| 00190 | 6.761e-03 | 0.2714 | 4 | 30 | Oxidative phosphorylation |
ACYP2acylphosphatase 2, muscle type (ENSG00000170634), score: -0.68 ARSKarylsulfatase family, member K (ENSG00000164291), score: 0.64 ATG4CATG4 autophagy related 4 homolog C (S. cerevisiae) (ENSG00000125703), score: 0.7 ATP2B4ATPase, Ca++ transporting, plasma membrane 4 (ENSG00000058668), score: -0.69 BLVRAbiliverdin reductase A (ENSG00000106605), score: -0.67 C11orf46chromosome 11 open reading frame 46 (ENSG00000152219), score: 0.67 C12orf66chromosome 12 open reading frame 66 (ENSG00000174206), score: 0.64 C13orf31chromosome 13 open reading frame 31 (ENSG00000179630), score: 0.7 C20orf43chromosome 20 open reading frame 43 (ENSG00000022277), score: -0.66 C3orf39chromosome 3 open reading frame 39 (ENSG00000144647), score: 0.66 CDC123cell division cycle 123 homolog (S. cerevisiae) (ENSG00000151465), score: -0.67 CDCA3cell division cycle associated 3 (ENSG00000111665), score: 0.77 COX1cytochrome c oxidase subunit I (ENSG00000198804), score: -0.95 CYTBcytochrome b (ENSG00000198727), score: -1 CYTH1cytohesin 1 (ENSG00000108669), score: -0.67 FAM73Afamily with sequence similarity 73, member A (ENSG00000180488), score: 0.67 FUT10fucosyltransferase 10 (alpha (1,3) fucosyltransferase) (ENSG00000172728), score: 0.8 IKIK cytokine, down-regulator of HLA II (ENSG00000113141), score: -0.74 IPPintracisternal A particle-promoted polypeptide (ENSG00000197429), score: 0.88 MEMO1mediator of cell motility 1 (ENSG00000162961), score: -0.65 METRNLmeteorin, glial cell differentiation regulator-like (ENSG00000176845), score: -0.67 NARFnuclear prelamin A recognition factor (ENSG00000141562), score: -0.69 ND2MTND2 (ENSG00000198763), score: -0.97 ND6NADH dehydrogenase, subunit 6 (complex I) (ENSG00000198695), score: -0.67 NIF3L1NIF3 NGG1 interacting factor 3-like 1 (S. pombe) (ENSG00000196290), score: 0.68 NOXO1NADPH oxidase organizer 1 (ENSG00000196408), score: 0.9 NSUN2NOP2/Sun domain family, member 2 (ENSG00000037474), score: -0.69 PDE6Gphosphodiesterase 6G, cGMP-specific, rod, gamma (ENSG00000185527), score: 0.64 PIGKphosphatidylinositol glycan anchor biosynthesis, class K (ENSG00000142892), score: 0.66 PIGLphosphatidylinositol glycan anchor biosynthesis, class L (ENSG00000108474), score: 0.76 PYCRLpyrroline-5-carboxylate reductase-like (ENSG00000104524), score: 0.75 RIC8Bresistance to inhibitors of cholinesterase 8 homolog B (C. elegans) (ENSG00000111785), score: 0.75 RPS6KA6ribosomal protein S6 kinase, 90kDa, polypeptide 6 (ENSG00000072133), score: 0.7 SLC38A8solute carrier family 38, member 8 (ENSG00000166558), score: 0.91 SNX13sorting nexin 13 (ENSG00000071189), score: 0.65 STT3BSTT3, subunit of the oligosaccharyltransferase complex, homolog B (S. cerevisiae) (ENSG00000163527), score: 0.68 THYN1thymocyte nuclear protein 1 (ENSG00000151500), score: -0.72 TMEM184Ctransmembrane protein 184C (ENSG00000164168), score: 0.81 TOE1target of EGR1, member 1 (nuclear) (ENSG00000132773), score: 0.71 UBE2G1ubiquitin-conjugating enzyme E2G 1 (UBC7 homolog, yeast) (ENSG00000132388), score: -0.8 ZC3H15zinc finger CCCH-type containing 15 (ENSG00000065548), score: -0.9 ZNF512zinc finger protein 512 (ENSG00000243943), score: 0.77 ZWILCHZwilch, kinetochore associated, homolog (Drosophila) (ENSG00000174442), score: 0.69
| Id | species | tissue | sex | individual |
|---|---|---|---|---|
| ggo_ht_m_ca1 | ggo | ht | m | _ |
| ggo_ht_f_ca1 | ggo | ht | f | _ |
| mdo_lv_f_ca1 | mdo | lv | f | _ |
| mdo_ht_m_ca1 | mdo | ht | m | _ |
| mdo_lv_m_ca1 | mdo | lv | m | _ |
| mdo_ts_m2_ca1 | mdo | ts | m | 2 |
| mdo_ts_m1_ca1 | mdo | ts | m | 1 |
| mdo_kd_m_ca1 | mdo | kd | m | _ |
| mdo_br_f_ca1 | mdo | br | f | _ |
| mdo_kd_f_ca1 | mdo | kd | f | _ |
| mdo_br_m_ca1 | mdo | br | m | _ |
| mdo_ht_f_ca1 | mdo | ht | f | _ |
| mdo_cb_f_ca1 | mdo | cb | f | _ |
| mdo_cb_m_ca1 | mdo | cb | m | _ |