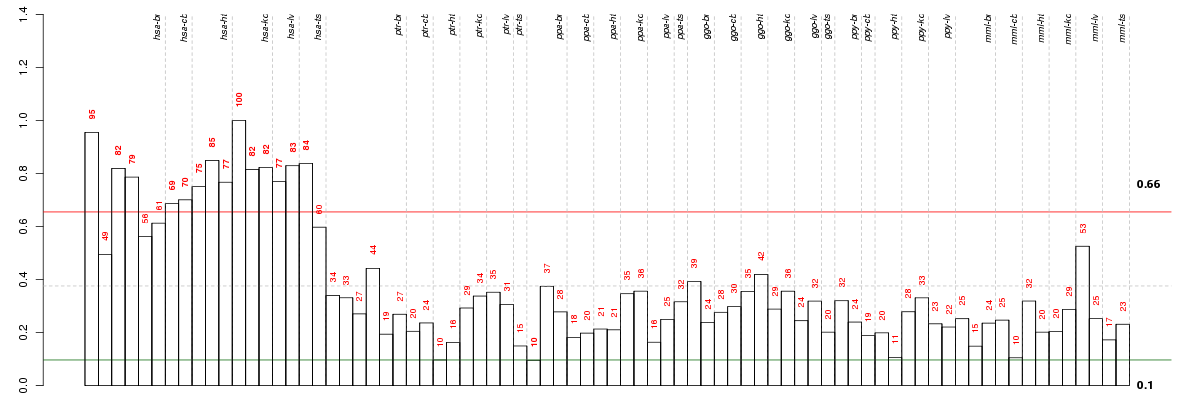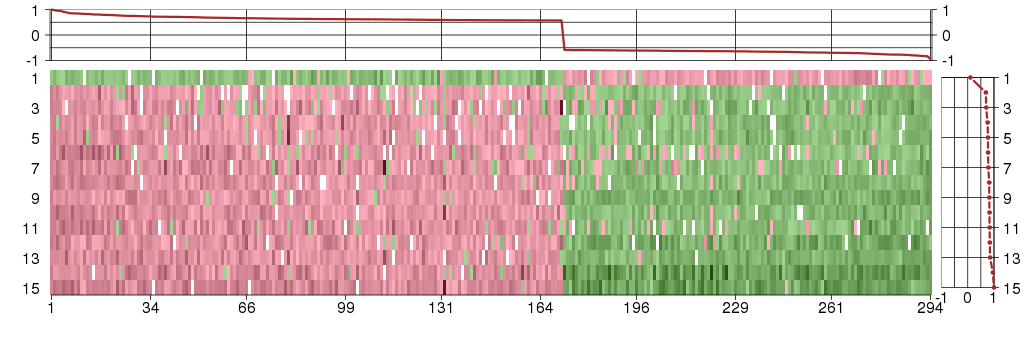

Under-expression is coded with green,
over-expression with red color.

metabolic process
The chemical reactions and pathways, including anabolism and catabolism, by which living organisms transform chemical substances. Metabolic processes typically transform small molecules, but also include macromolecular processes such as DNA repair and replication, and protein synthesis and degradation.
nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
Any cellular metabolic process involving nucleobases, nucleosides, nucleotides and nucleic acids.
RNA processing
Any process involved in the conversion of one or more primary RNA transcripts into one or more mature RNA molecules.
mRNA processing
Any process involved in the conversion of a primary mRNA transcript into one or more mature mRNA(s) prior to translation into polypeptide.
nitrogen compound metabolic process
The chemical reactions and pathways involving various organic and inorganic nitrogenous compounds; includes nitrogen fixation, nitrification, denitrification, assimilatory/dissimilatory nitrate reduction and the interconversion of nitrogenous organic matter and ammonium.
organelle organization
A process that is carried out at the cellular level which results in the assembly, arrangement of constituent parts, or disassembly of an organelle within a cell. An organelle is an organized structure of distinctive morphology and function. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton. Excludes the plasma membrane.
mitochondrion organization
A process that is carried out at the cellular level which results in the assembly, arrangement of constituent parts, or disassembly of a mitochondrion; includes mitochondrial morphogenesis and distribution, and replication of the mitochondrial genome as well as synthesis of new mitochondrial components.
biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
RNA splicing
The process of removing sections of the primary RNA transcript to remove sequences not present in the mature form of the RNA and joining the remaining sections to form the mature form of the RNA.
catabolic process
The chemical reactions and pathways resulting in the breakdown of substances, including the breakdown of carbon compounds with the liberation of energy for use by the cell or organism.
macromolecule catabolic process
The chemical reactions and pathways resulting in the breakdown of a macromolecule, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass.
cellular process
Any process that is carried out at the cellular level, but not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
gene expression
The process by which a gene's sequence is converted into a mature gene product or products (proteins or RNA). This includes the production of an RNA transcript as well as any processing to produce a mature RNA product or an mRNA (for protein-coding genes) and the translation of that mRNA into protein. Some protein processing events may be included when they are required to form an active form of a product from an inactive precursor form.
cellular component organization
A process that is carried out at the cellular level which results in the assembly, arrangement of constituent parts, or disassembly of a cellular component.
RNA metabolic process
The cellular chemical reactions and pathways involving RNA, ribonucleic acid, one of the two main type of nucleic acid, consisting of a long, unbranched macromolecule formed from ribonucleotides joined in 3',5'-phosphodiester linkage.
mRNA metabolic process
The chemical reactions and pathways involving mRNA, messenger RNA, which is responsible for carrying the coded genetic 'message', transcribed from DNA, to sites of protein assembly at the ribosomes.
cellular nitrogen compound metabolic process
The chemical reactions and pathways involving various organic and inorganic nitrogenous compounds, as carried out by individual cells.
macromolecule metabolic process
The chemical reactions and pathways involving macromolecules, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass.
cellular metabolic process
The chemical reactions and pathways by which individual cells transform chemical substances.
primary metabolic process
The chemical reactions and pathways involving those compounds which are formed as a part of the normal anabolic and catabolic processes. These processes take place in most, if not all, cells of the organism.
cellular catabolic process
The chemical reactions and pathways resulting in the breakdown of substances, carried out by individual cells.
cellular macromolecule metabolic process
The chemical reactions and pathways involving macromolecules, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass, as carried out by individual cells.
cellular macromolecule catabolic process
The chemical reactions and pathways resulting in the breakdown of a macromolecule, any large molecule including proteins, nucleic acids and carbohydrates, as carried out by individual cells.
nucleic acid metabolic process
Any cellular metabolic process involving nucleic acids.
all
NA
cellular metabolic process
The chemical reactions and pathways by which individual cells transform chemical substances.
organelle organization
A process that is carried out at the cellular level which results in the assembly, arrangement of constituent parts, or disassembly of an organelle within a cell. An organelle is an organized structure of distinctive morphology and function. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton. Excludes the plasma membrane.
macromolecule catabolic process
The chemical reactions and pathways resulting in the breakdown of a macromolecule, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass.
cellular nitrogen compound metabolic process
The chemical reactions and pathways involving various organic and inorganic nitrogenous compounds, as carried out by individual cells.
cellular catabolic process
The chemical reactions and pathways resulting in the breakdown of substances, carried out by individual cells.
cellular macromolecule metabolic process
The chemical reactions and pathways involving macromolecules, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass, as carried out by individual cells.
nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
Any cellular metabolic process involving nucleobases, nucleosides, nucleotides and nucleic acids.
cellular macromolecule catabolic process
The chemical reactions and pathways resulting in the breakdown of a macromolecule, any large molecule including proteins, nucleic acids and carbohydrates, as carried out by individual cells.
cellular macromolecule catabolic process
The chemical reactions and pathways resulting in the breakdown of a macromolecule, any large molecule including proteins, nucleic acids and carbohydrates, as carried out by individual cells.
RNA processing
Any process involved in the conversion of one or more primary RNA transcripts into one or more mature RNA molecules.
RNA metabolic process
The cellular chemical reactions and pathways involving RNA, ribonucleic acid, one of the two main type of nucleic acid, consisting of a long, unbranched macromolecule formed from ribonucleotides joined in 3',5'-phosphodiester linkage.
mRNA processing
Any process involved in the conversion of a primary mRNA transcript into one or more mature mRNA(s) prior to translation into polypeptide.

ubiquitin ligase complex
A protein complex that includes a ubiquitin-protein ligase and other proteins that may confer substrate specificity on the complex.
intracellular
The living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
ribonucleoprotein complex
A macromolecular complex containing both protein and RNA molecules.
small nuclear ribonucleoprotein complex
A complex composed of RNA of the small nuclear RNA (snRNA) class and protein, found in the nucleus of a eukaryotic cell. These are typically named after the snRNA(s) they contain, e.g. U1 snRNP or U4/U6 snRNP. Many, but not all, of these complexes are involved in splicing of nuclear mRNAs.
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
cell
The basic structural and functional unit of all organisms. Includes the plasma membrane and any external encapsulating structures such as the cell wall and cell envelope.
nucleus
A membrane-bounded organelle of eukaryotic cells in which chromosomes are housed and replicated. In most cells, the nucleus contains all of the cell's chromosomes except the organellar chromosomes, and is the site of RNA synthesis and processing. In some species, or in specialized cell types, RNA metabolism or DNA replication may be absent.
spliceosomal complex
Any of a series of ribonucleoprotein complexes that contain RNA and small nuclear ribonucleoproteins (snRNPs), and are formed sequentially during the splicing of a messenger RNA primary transcript to excise an intron.
U4 snRNP
A ribonucleoprotein complex that contains small nuclear RNA U4.
nucleolus
A small, dense body one or more of which are present in the nucleus of eukaryotic cells. It is rich in RNA and protein, is not bounded by a limiting membrane, and is not seen during mitosis. Its prime function is the transcription of the nucleolar DNA into 45S ribosomal-precursor RNA, the processing of this RNA into 5.8S, 18S, and 28S components of ribosomal RNA, and the association of these components with 5S RNA and proteins synthesized outside the nucleolus. This association results in the formation of ribonucleoprotein precursors; these pass into the cytoplasm and mature into the 40S and 60S subunits of the ribosome.
cullin-RING ubiquitin ligase complex
Any ubiquitin ligase complex in which the catalytic core consists of a member of the cullin family and a RING domain protein; the core is associated with one or more additional proteins that confer substrate specificity.
Cul4A-RING ubiquitin ligase complex
A ubiquitin ligase complex in which a cullin from the Cul4A subfamily and a RING domain protein form the catalytic core; substrate specificity is conferred by an adaptor protein.
membrane-enclosed lumen
The enclosed volume within a sealed membrane or between two sealed membranes. Encompasses the volume enclosed by the membranes of a particular organelle, e.g. endoplasmic reticulum lumen, or the space between the two lipid bilayers of a double membrane surrounding an organelle, e.g. nuclear envelope lumen.
nuclear lumen
The volume enclosed by the nuclear inner membrane.
macromolecular complex
A stable assembly of two or more macromolecules, i.e. proteins, nucleic acids, carbohydrates or lipids, in which the constituent parts function together.
organelle
Organized structure of distinctive morphology and function. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton, and prokaryotic structures such as anammoxosomes and pirellulosomes. Excludes the plasma membrane.
membrane-bounded organelle
Organized structure of distinctive morphology and function, bounded by a single or double lipid bilayer membrane. Includes the nucleus, mitochondria, plastids, vacuoles, and vesicles. Excludes the plasma membrane.
non-membrane-bounded organelle
Organized structure of distinctive morphology and function, not bounded by a lipid bilayer membrane. Includes ribosomes, the cytoskeleton and chromosomes.
intracellular organelle
Organized structure of distinctive morphology and function, occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton. Excludes the plasma membrane.
intracellular membrane-bounded organelle
Organized structure of distinctive morphology and function, bounded by a single or double lipid bilayer membrane and occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, and vesicles. Excludes the plasma membrane.
intracellular non-membrane-bounded organelle
Organized structure of distinctive morphology and function, not bounded by a lipid bilayer membrane and occurring within the cell. Includes ribosomes, the cytoskeleton and chromosomes.
organelle lumen
The internal volume enclosed by the membranes of a particular organelle; includes the volume enclosed by a single organelle membrane, e.g. endoplasmic reticulum lumen, or the volume enclosed by the innermost of the two lipid bilayers of an organelle envelope, e.g. nuclear lumen.
protein complex
Any macromolecular complex composed of two or more polypeptide subunits, which may or may not be identical. Protein complexes may have other associated non-protein prosthetic groups, such as nucleotides, metal ions or other small molecules.
organelle part
Any constituent part of an organelle, an organized structure of distinctive morphology and function. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton, but excludes the plasma membrane.
intracellular part
Any constituent part of the living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
nuclear part
Any constituent part of the nucleus, a membrane-bounded organelle of eukaryotic cells in which chromosomes are housed and replicated.
intracellular organelle part
A constituent part of an intracellular organelle, an organized structure of distinctive morphology and function, occurring within the cell. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton but excludes the plasma membrane.
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
intracellular organelle lumen
An organelle lumen that is part of an intracellular organelle.
CUL4 RING ubiquitin ligase complex
A ubiquitin ligase complex in which a cullin from the Cul4 family and a RING domain protein form the catalytic core; substrate specificity is conferred by an adaptor protein.
all
NA
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
organelle part
Any constituent part of an organelle, an organized structure of distinctive morphology and function. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton, but excludes the plasma membrane.
organelle lumen
The internal volume enclosed by the membranes of a particular organelle; includes the volume enclosed by a single organelle membrane, e.g. endoplasmic reticulum lumen, or the volume enclosed by the innermost of the two lipid bilayers of an organelle envelope, e.g. nuclear lumen.
intracellular membrane-bounded organelle
Organized structure of distinctive morphology and function, bounded by a single or double lipid bilayer membrane and occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, and vesicles. Excludes the plasma membrane.
intracellular non-membrane-bounded organelle
Organized structure of distinctive morphology and function, not bounded by a lipid bilayer membrane and occurring within the cell. Includes ribosomes, the cytoskeleton and chromosomes.
intracellular organelle part
A constituent part of an intracellular organelle, an organized structure of distinctive morphology and function, occurring within the cell. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton but excludes the plasma membrane.
intracellular organelle lumen
An organelle lumen that is part of an intracellular organelle.
intracellular part
Any constituent part of the living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
ubiquitin ligase complex
A protein complex that includes a ubiquitin-protein ligase and other proteins that may confer substrate specificity on the complex.
ribonucleoprotein complex
A macromolecular complex containing both protein and RNA molecules.
intracellular organelle
Organized structure of distinctive morphology and function, occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton. Excludes the plasma membrane.
intracellular organelle part
A constituent part of an intracellular organelle, an organized structure of distinctive morphology and function, occurring within the cell. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton but excludes the plasma membrane.
small nuclear ribonucleoprotein complex
A complex composed of RNA of the small nuclear RNA (snRNA) class and protein, found in the nucleus of a eukaryotic cell. These are typically named after the snRNA(s) they contain, e.g. U1 snRNP or U4/U6 snRNP. Many, but not all, of these complexes are involved in splicing of nuclear mRNAs.
spliceosomal complex
Any of a series of ribonucleoprotein complexes that contain RNA and small nuclear ribonucleoproteins (snRNPs), and are formed sequentially during the splicing of a messenger RNA primary transcript to excise an intron.
nucleolus
A small, dense body one or more of which are present in the nucleus of eukaryotic cells. It is rich in RNA and protein, is not bounded by a limiting membrane, and is not seen during mitosis. Its prime function is the transcription of the nucleolar DNA into 45S ribosomal-precursor RNA, the processing of this RNA into 5.8S, 18S, and 28S components of ribosomal RNA, and the association of these components with 5S RNA and proteins synthesized outside the nucleolus. This association results in the formation of ribonucleoprotein precursors; these pass into the cytoplasm and mature into the 40S and 60S subunits of the ribosome.
nuclear lumen
The volume enclosed by the nuclear inner membrane.
nucleolus
A small, dense body one or more of which are present in the nucleus of eukaryotic cells. It is rich in RNA and protein, is not bounded by a limiting membrane, and is not seen during mitosis. Its prime function is the transcription of the nucleolar DNA into 45S ribosomal-precursor RNA, the processing of this RNA into 5.8S, 18S, and 28S components of ribosomal RNA, and the association of these components with 5S RNA and proteins synthesized outside the nucleolus. This association results in the formation of ribonucleoprotein precursors; these pass into the cytoplasm and mature into the 40S and 60S subunits of the ribosome.
nuclear part
Any constituent part of the nucleus, a membrane-bounded organelle of eukaryotic cells in which chromosomes are housed and replicated.

molecular_function
Elemental activities, such as catalysis or binding, describing the actions of a gene product at the molecular level. A given gene product may exhibit one or more molecular functions.
nucleic acid binding
Interacting selectively and non-covalently with any nucleic acid.
RNA binding
Interacting selectively and non-covalently with an RNA molecule or a portion thereof.
binding
The selective, non-covalent, often stoichiometric, interaction of a molecule with one or more specific sites on another molecule.
all
NA
| Id | Pvalue | ExpCount | Count | Size | Term |
|---|---|---|---|---|---|
| 03040 | 5.234e-03 | 1.64 | 8 | 85 | Spliceosome |
| 04120 | 1.637e-02 | 2.006 | 8 | 104 | Ubiquitin mediated proteolysis |
AAMPangio-associated, migratory cell protein (ENSG00000127837), score: 0.58 ABCE1ATP-binding cassette, sub-family E (OABP), member 1 (ENSG00000164163), score: -0.68 ABHD13abhydrolase domain containing 13 (ENSG00000139826), score: -0.61 ACAP2ArfGAP with coiled-coil, ankyrin repeat and PH domains 2 (ENSG00000114331), score: -0.65 ACOT8acyl-CoA thioesterase 8 (ENSG00000101473), score: 0.7 AGXT2L2alanine-glyoxylate aminotransferase 2-like 2 (ENSG00000175309), score: 0.57 AIDAaxin interactor, dorsalization associated (ENSG00000186063), score: -0.74 AIParyl hydrocarbon receptor interacting protein (ENSG00000110711), score: 0.63 AKT1S1AKT1 substrate 1 (proline-rich) (ENSG00000204673), score: 0.61 AKTIPAKT interacting protein (ENSG00000166971), score: -0.7 ANAPC4anaphase promoting complex subunit 4 (ENSG00000053900), score: -0.66 AP4M1adaptor-related protein complex 4, mu 1 subunit (ENSG00000221838), score: 0.62 AS3MTarsenic (+3 oxidation state) methyltransferase (ENSG00000214435), score: 0.59 ATG10ATG10 autophagy related 10 homolog (S. cerevisiae) (ENSG00000152348), score: 0.77 ATG12ATG12 autophagy related 12 homolog (S. cerevisiae) (ENSG00000145782), score: -0.71 ATG4CATG4 autophagy related 4 homolog C (S. cerevisiae) (ENSG00000125703), score: -0.62 BAT1HLA-B associated transcript 1 (ENSG00000198563), score: 0.59 BAT5HLA-B associated transcript 5 (ENSG00000204422), score: 1 BDP1B double prime 1, subunit of RNA polymerase III transcription initiation factor IIIB (ENSG00000145734), score: -0.63 C11orf17chromosome 11 open reading frame 17 (ENSG00000166452), score: 0.61 C11orf2chromosome 11 open reading frame 2 (ENSG00000149823), score: 0.68 C12orf4chromosome 12 open reading frame 4 (ENSG00000047621), score: -0.72 C12orf52chromosome 12 open reading frame 52 (ENSG00000139405), score: 0.67 C14orf80chromosome 14 open reading frame 80 (ENSG00000185347), score: 0.74 C15orf29chromosome 15 open reading frame 29 (ENSG00000134152), score: -0.59 C17orf103chromosome 17 open reading frame 103 (ENSG00000154035), score: 0.57 C1orf107chromosome 1 open reading frame 107 (ENSG00000117597), score: -0.63 C1orf186chromosome 1 open reading frame 186 (ENSG00000196533), score: 0.59 C1orf43chromosome 1 open reading frame 43 (ENSG00000143612), score: 0.58 C2orf24chromosome 2 open reading frame 24 (ENSG00000115649), score: 0.57 C2orf49chromosome 2 open reading frame 49 (ENSG00000135974), score: -0.77 C2orf79chromosome 2 open reading frame 79 (ENSG00000184924), score: 0.63 C3orf16chromosome 3 open reading frame 16 (ENSG00000206199), score: 0.75 C5orf15chromosome 5 open reading frame 15 (ENSG00000113583), score: 0.67 C5orf41chromosome 5 open reading frame 41 (ENSG00000164463), score: -0.71 C5orf44chromosome 5 open reading frame 44 (ENSG00000113597), score: -0.65 C5orf45chromosome 5 open reading frame 45 (ENSG00000161010), score: 0.84 C6orf162chromosome 6 open reading frame 162 (ENSG00000111850), score: -0.69 C6orf182chromosome 6 open reading frame 182 (ENSG00000183137), score: -0.63 C6orf62chromosome 6 open reading frame 62 (ENSG00000112308), score: -0.66 C8orf37chromosome 8 open reading frame 37 (ENSG00000156172), score: -0.66 C8orf38chromosome 8 open reading frame 38 (ENSG00000156170), score: -0.7 C9orf142chromosome 9 open reading frame 142 (ENSG00000148362), score: 0.61 CCNB1IP1cyclin B1 interacting protein 1 (ENSG00000100814), score: 0.58 CCNT2cyclin T2 (ENSG00000082258), score: -0.69 CCPG1cell cycle progression 1 (ENSG00000214882), score: -0.61 CD72CD72 molecule (ENSG00000137101), score: 0.79 CDC26cell division cycle 26 homolog (S. cerevisiae) (ENSG00000176386), score: -0.75 CDC27cell division cycle 27 homolog (S. cerevisiae) (ENSG00000004897), score: -0.7 CECR5cat eye syndrome chromosome region, candidate 5 (ENSG00000069998), score: 0.79 CENPTcentromere protein T (ENSG00000102901), score: 0.61 CEP290centrosomal protein 290kDa (ENSG00000198707), score: -0.59 CIITAclass II, major histocompatibility complex, transactivator (ENSG00000179583), score: 0.8 CISD2CDGSH iron sulfur domain 2 (ENSG00000145354), score: 0.61 CITED4Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 4 (ENSG00000179862), score: 0.73 CLCC1chloride channel CLIC-like 1 (ENSG00000121940), score: -0.7 CLN8ceroid-lipofuscinosis, neuronal 8 (epilepsy, progressive with mental retardation) (ENSG00000182372), score: 0.72 CLPPClpP caseinolytic peptidase, ATP-dependent, proteolytic subunit homolog (E. coli) (ENSG00000125656), score: 0.61 CLUL1clusterin-like 1 (retinal) (ENSG00000079101), score: 0.57 CNIH4cornichon homolog 4 (Drosophila) (ENSG00000143771), score: -0.63 COASYCoA synthase (ENSG00000068120), score: 0.67 COMMD2COMM domain containing 2 (ENSG00000114744), score: -0.8 COQ4coenzyme Q4 homolog (S. cerevisiae) (ENSG00000167113), score: 0.63 COX4NBCOX4 neighbor (ENSG00000131148), score: 0.63 CRBNcereblon (ENSG00000113851), score: -0.73 CRIPTcysteine-rich PDZ-binding protein (ENSG00000119878), score: -0.76 CWC22CWC22 spliceosome-associated protein homolog (S. cerevisiae) (ENSG00000163510), score: -0.6 CWF19L2CWF19-like 2, cell cycle control (S. pombe) (ENSG00000152404), score: -0.64 CYB561D2cytochrome b-561 domain containing 2 (ENSG00000114395), score: 0.63 CYLDcylindromatosis (turban tumor syndrome) (ENSG00000083799), score: -0.63 CYP4B1cytochrome P450, family 4, subfamily B, polypeptide 1 (ENSG00000142973), score: 0.64 DDX5DEAD (Asp-Glu-Ala-Asp) box polypeptide 5 (ENSG00000108654), score: -0.6 DDX54DEAD (Asp-Glu-Ala-Asp) box polypeptide 54 (ENSG00000123064), score: 0.58 DHPSdeoxyhypusine synthase (ENSG00000095059), score: 0.61 DHX38DEAH (Asp-Glu-Ala-His) box polypeptide 38 (ENSG00000140829), score: 0.59 DLEC1deleted in lung and esophageal cancer 1 (ENSG00000008226), score: 0.79 DNLZDNL-type zinc finger (ENSG00000213221), score: 0.57 DOHHdeoxyhypusine hydroxylase/monooxygenase (ENSG00000129932), score: 0.63 DPH1DPH1 homolog (S. cerevisiae) (ENSG00000108963), score: 0.72 DUS4Ldihydrouridine synthase 4-like (S. cerevisiae) (ENSG00000105865), score: -0.64 DUSP11dual specificity phosphatase 11 (RNA/RNP complex 1-interacting) (ENSG00000144048), score: -0.69 EEA1early endosome antigen 1 (ENSG00000102189), score: -0.69 EEF1E1eukaryotic translation elongation factor 1 epsilon 1 (ENSG00000124802), score: -0.64 EEF1Geukaryotic translation elongation factor 1 gamma (ENSG00000149016), score: 0.97 EEF2eukaryotic translation elongation factor 2 (ENSG00000167658), score: 0.74 EIF3Keukaryotic translation initiation factor 3, subunit K (ENSG00000178982), score: 0.7 ELAC2elaC homolog 2 (E. coli) (ENSG00000006744), score: 0.89 ERCC6excision repair cross-complementing rodent repair deficiency, complementation group 6 (ENSG00000225830), score: -0.66 ERCC8excision repair cross-complementing rodent repair deficiency, complementation group 8 (ENSG00000049167), score: -0.64 ERI3ERI1 exoribonuclease family member 3 (ENSG00000117419), score: 0.6 ESF1ESF1, nucleolar pre-rRNA processing protein, homolog (S. cerevisiae) (ENSG00000089048), score: -0.62 EXOC1exocyst complex component 1 (ENSG00000090989), score: -0.73 EXOC6exocyst complex component 6 (ENSG00000138190), score: -0.61 EXOGendo/exonuclease (5'-3'), endonuclease G-like (ENSG00000157036), score: -0.61 FABP6fatty acid binding protein 6, ileal (ENSG00000170231), score: 0.62 FAM103A1family with sequence similarity 103, member A1 (ENSG00000169612), score: -0.84 FAM113Afamily with sequence similarity 113, member A (ENSG00000132635), score: 0.58 FAM119Afamily with sequence similarity 119, member A (ENSG00000144401), score: -0.69 FAM156Afamily with sequence similarity 156, member A (ENSG00000182646), score: -0.82 FAM175Bfamily with sequence similarity 175, member B (ENSG00000165660), score: -0.59 FAM63Afamily with sequence similarity 63, member A (ENSG00000143409), score: 0.63 FNTBfarnesyltransferase, CAAX box, beta (ENSG00000125954), score: 0.62 FOXP4forkhead box P4 (ENSG00000137166), score: 0.58 FRA10AC1fragile site, folic acid type, rare, fra(10)(q23.3) or fra(10)(q24.2) candidate 1 (ENSG00000148690), score: -0.7 GIPC1GIPC PDZ domain containing family, member 1 (ENSG00000123159), score: 0.65 GIPRgastric inhibitory polypeptide receptor (ENSG00000010310), score: 0.61 GLT25D1glycosyltransferase 25 domain containing 1 (ENSG00000130309), score: 0.63 GMPPBGDP-mannose pyrophosphorylase B (ENSG00000173540), score: 0.57 GNB2guanine nucleotide binding protein (G protein), beta polypeptide 2 (ENSG00000172354), score: 0.61 GNPTGN-acetylglucosamine-1-phosphate transferase, gamma subunit (ENSG00000090581), score: 0.58 GORABgolgin, RAB6-interacting (ENSG00000120370), score: -0.6 GPN2GPN-loop GTPase 2 (ENSG00000142751), score: 0.63 GPR141G protein-coupled receptor 141 (ENSG00000187037), score: 0.75 GPR44G protein-coupled receptor 44 (ENSG00000183134), score: 0.63 GPS1G protein pathway suppressor 1 (ENSG00000169727), score: 0.66 GRWD1glutamate-rich WD repeat containing 1 (ENSG00000105447), score: 0.71 GSPT1G1 to S phase transition 1 (ENSG00000103342), score: -0.67 GUK1guanylate kinase 1 (ENSG00000143774), score: 0.75 HECTD2HECT domain containing 2 (ENSG00000165338), score: -0.59 HERC4hect domain and RLD 4 (ENSG00000148634), score: -0.68 HMBOX1homeobox containing 1 (ENSG00000147421), score: -0.6 HOOK3hook homolog 3 (Drosophila) (ENSG00000168172), score: -0.66 HSPA1Lheat shock 70kDa protein 1-like (ENSG00000204390), score: -0.61 HTRA2HtrA serine peptidase 2 (ENSG00000115317), score: 0.62 IFT27intraflagellar transport 27 homolog (Chlamydomonas) (ENSG00000100360), score: 0.58 IL18BPinterleukin 18 binding protein (ENSG00000137496), score: 0.68 IL9Rinterleukin 9 receptor (ENSG00000124334), score: 0.95 ILVBLilvB (bacterial acetolactate synthase)-like (ENSG00000105135), score: 0.58 INMTindolethylamine N-methyltransferase (ENSG00000106125), score: 0.77 INTS2integrator complex subunit 2 (ENSG00000108506), score: -0.65 ITGAEintegrin, alpha E (antigen CD103, human mucosal lymphocyte antigen 1; alpha polypeptide) (ENSG00000083457), score: 0.59 ITGB3BPintegrin beta 3 binding protein (beta3-endonexin) (ENSG00000142856), score: 0.65 KEAP1kelch-like ECH-associated protein 1 (ENSG00000079999), score: 0.57 KIAA1715KIAA1715 (ENSG00000144320), score: -0.62 KINKIN, antigenic determinant of recA protein homolog (mouse) (ENSG00000151657), score: -0.66 KRR1KRR1, small subunit (SSU) processome component, homolog (yeast) (ENSG00000111615), score: -0.62 LCN12lipocalin 12 (ENSG00000184925), score: 0.66 LGALS3BPlectin, galactoside-binding, soluble, 3 binding protein (ENSG00000108679), score: 0.67 LOC100131693eukaryotic translation initiation factor 4E pseudogene (ENSG00000151247), score: -0.7 LOC100133770hypothetical protein LOC100133770 (ENSG00000069329), score: -0.74 LOC652522similar to Werner syndrome protein (ENSG00000165392), score: -0.69 LOC731605similar to Bcl-2-associated transcription factor 1 (Btf) (ENSG00000029363), score: -0.72 LONP1lon peptidase 1, mitochondrial (ENSG00000196365), score: 0.57 LPAR6lysophosphatidic acid receptor 6 (ENSG00000139679), score: -0.63 LRSAM1leucine rich repeat and sterile alpha motif containing 1 (ENSG00000148356), score: 0.57 LSM12LSM12 homolog (S. cerevisiae) (ENSG00000161654), score: -0.59 LTBP3latent transforming growth factor beta binding protein 3 (ENSG00000168056), score: 0.57 MAEAmacrophage erythroblast attacher (ENSG00000090316), score: 0.58 MAK16MAK16 homolog (S. cerevisiae) (ENSG00000198042), score: -0.64 MAN2B2mannosidase, alpha, class 2B, member 2 (ENSG00000013288), score: 0.6 MAPKAPK5mitogen-activated protein kinase-activated protein kinase 5 (ENSG00000089022), score: 0.62 MED16mediator complex subunit 16 (ENSG00000175221), score: 0.64 METT11D1methyltransferase 11 domain containing 1 (ENSG00000165792), score: 0.61 METTL6methyltransferase like 6 (ENSG00000206562), score: -0.66 MFN1mitofusin 1 (ENSG00000171109), score: -0.6 MFSD7major facilitator superfamily domain containing 7 (ENSG00000169026), score: 0.61 MGRN1mahogunin, ring finger 1 (ENSG00000102858), score: 0.63 MIFmacrophage migration inhibitory factor (glycosylation-inhibiting factor) (ENSG00000133460), score: 0.84 MLST8MTOR associated protein, LST8 homolog (S. cerevisiae) (ENSG00000167965), score: 0.66 MMABmethylmalonic aciduria (cobalamin deficiency) cblB type (ENSG00000139428), score: 0.6 MNAT1menage a trois homolog 1, cyclin H assembly factor (Xenopus laevis) (ENSG00000020426), score: -0.67 MOBKL2AMOB1, Mps One Binder kinase activator-like 2A (yeast) (ENSG00000172081), score: 0.62 MPHOSPH10M-phase phosphoprotein 10 (U3 small nucleolar ribonucleoprotein) (ENSG00000124383), score: -0.59 MTA2metastasis associated 1 family, member 2 (ENSG00000149480), score: 0.64 MYL5myosin, light chain 5, regulatory (ENSG00000215375), score: 0.85 NAA25N(alpha)-acetyltransferase 25, NatB auxiliary subunit (ENSG00000111300), score: -0.61 NDOR1NADPH dependent diflavin oxidoreductase 1 (ENSG00000188566), score: 0.68 NFICnuclear factor I/C (CCAAT-binding transcription factor) (ENSG00000141905), score: 0.59 NOL8nucleolar protein 8 (ENSG00000198000), score: -0.77 NPBneuropeptide B (ENSG00000183979), score: 0.82 NPRL3nitrogen permease regulator-like 3 (S. cerevisiae) (ENSG00000103148), score: 0.73 NSFL1CNSFL1 (p97) cofactor (p47) (ENSG00000088833), score: 0.57 NT5C35'-nucleotidase, cytosolic III (ENSG00000122643), score: -0.66 NT5DC25'-nucleotidase domain containing 2 (ENSG00000168268), score: 0.71 ODF2Louter dense fiber of sperm tails 2-like (ENSG00000122417), score: -0.77 OTUD6BOTU domain containing 6B (ENSG00000155100), score: -0.7 PABPC1Lpoly(A) binding protein, cytoplasmic 1-like (ENSG00000101104), score: 0.69 PAPOLGpoly(A) polymerase gamma (ENSG00000115421), score: -0.59 PEX10peroxisomal biogenesis factor 10 (ENSG00000157911), score: 0.7 PFKLphosphofructokinase, liver (ENSG00000141959), score: 0.59 PHF14PHD finger protein 14 (ENSG00000106443), score: -0.71 PIGCphosphatidylinositol glycan anchor biosynthesis, class C (ENSG00000135845), score: -0.78 PIGQphosphatidylinositol glycan anchor biosynthesis, class Q (ENSG00000007541), score: 0.58 PIGTphosphatidylinositol glycan anchor biosynthesis, class T (ENSG00000124155), score: 0.69 PIK3CDphosphoinositide-3-kinase, catalytic, delta polypeptide (ENSG00000171608), score: 0.65 PLEKHJ1pleckstrin homology domain containing, family J member 1 (ENSG00000104886), score: 0.65 PMS2PMS2 postmeiotic segregation increased 2 (S. cerevisiae) (ENSG00000122512), score: -0.61 PNPT1polyribonucleotide nucleotidyltransferase 1 (ENSG00000138035), score: -0.61 POLR2Lpolymerase (RNA) II (DNA directed) polypeptide L, 7.6kDa (ENSG00000177700), score: 0.61 POMGNT1protein O-linked mannose beta1,2-N-acetylglucosaminyltransferase (ENSG00000085998), score: 0.66 PPIGpeptidylprolyl isomerase G (cyclophilin G) (ENSG00000138398), score: -0.59 PRDX5peroxiredoxin 5 (ENSG00000126432), score: 0.58 PRICKLE4prickle homolog 4 (Drosophila) (ENSG00000124593), score: 0.68 PRPF31PRP31 pre-mRNA processing factor 31 homolog (S. cerevisiae) (ENSG00000105618), score: 0.62 PRPF39PRP39 pre-mRNA processing factor 39 homolog (S. cerevisiae) (ENSG00000185246), score: -0.62 PSAPprosaposin (ENSG00000197746), score: 0.72 PSMD9proteasome (prosome, macropain) 26S subunit, non-ATPase, 9 (ENSG00000110801), score: 0.65 PTCD2pentatricopeptide repeat domain 2 (ENSG00000049883), score: -0.63 RAB11BRAB11B, member RAS oncogene family (ENSG00000185236), score: 0.72 RABAC1Rab acceptor 1 (prenylated) (ENSG00000105404), score: 0.64 RBCK1RanBP-type and C3HC4-type zinc finger containing 1 (ENSG00000125826), score: 0.57 RBM8ARNA binding motif protein 8A (ENSG00000131795), score: -0.82 RPUSD4RNA pseudouridylate synthase domain containing 4 (ENSG00000165526), score: 0.62 RSPH9radial spoke head 9 homolog (Chlamydomonas) (ENSG00000172426), score: 0.6 RSPO1R-spondin homolog (Xenopus laevis) (ENSG00000169218), score: 0.83 RTF1Rtf1, Paf1/RNA polymerase II complex component, homolog (S. cerevisiae) (ENSG00000137815), score: -0.59 S100A13S100 calcium binding protein A13 (ENSG00000189171), score: 0.58 SCNN1Dsodium channel, nonvoltage-gated 1, delta (ENSG00000162572), score: 0.75 SCRIBscribbled homolog (Drosophila) (ENSG00000180900), score: 0.71 SDR39U1short chain dehydrogenase/reductase family 39U, member 1 (ENSG00000100445), score: 0.61 SETD6SET domain containing 6 (ENSG00000103037), score: -0.61 SFRS12splicing factor, arginine/serine-rich 12 (ENSG00000153914), score: -0.76 SFRS12IP1SFRS12-interacting protein 1 (ENSG00000153006), score: -0.63 SGTAsmall glutamine-rich tetratricopeptide repeat (TPR)-containing, alpha (ENSG00000104969), score: 0.59 SIVA1SIVA1, apoptosis-inducing factor (ENSG00000184990), score: 0.66 SLA2Src-like-adaptor 2 (ENSG00000101082), score: 0.68 SLC18A2solute carrier family 18 (vesicular monoamine), member 2 (ENSG00000165646), score: 0.84 SLC19A1solute carrier family 19 (folate transporter), member 1 (ENSG00000173638), score: 0.91 SNRPAsmall nuclear ribonucleoprotein polypeptide A (ENSG00000077312), score: 0.64 SNX13sorting nexin 13 (ENSG00000071189), score: -0.77 SNX17sorting nexin 17 (ENSG00000115234), score: 0.57 SNX22sorting nexin 22 (ENSG00000157734), score: 0.95 SR140U2-associated SR140 protein (ENSG00000163714), score: -0.7 SRMSsrc-related kinase lacking C-terminal regulatory tyrosine and N-terminal myristylation sites (ENSG00000125508), score: 0.57 SRSF11serine/arginine-rich splicing factor 11 (ENSG00000116754), score: -0.6 SSBSjogren syndrome antigen B (autoantigen La) (ENSG00000138385), score: -0.63 STRAPserine/threonine kinase receptor associated protein (ENSG00000023734), score: 0.58 STXBP3syntaxin binding protein 3 (ENSG00000116266), score: -0.62 SUN1Sad1 and UNC84 domain containing 1 (ENSG00000164828), score: 0.59 SURF6surfeit 6 (ENSG00000148296), score: 0.71 SUZ12suppressor of zeste 12 homolog (Drosophila) (ENSG00000178691), score: -0.68 TALDO1transaldolase 1 (ENSG00000177156), score: 0.6 TAS2R14taste receptor, type 2, member 14 (ENSG00000212127), score: 0.66 TBC1D8TBC1 domain family, member 8 (with GRAM domain) (ENSG00000204634), score: 0.78 TBK1TANK-binding kinase 1 (ENSG00000183735), score: -0.63 TH1LTH1-like (Drosophila) (ENSG00000101158), score: 0.81 THAP4THAP domain containing 4 (ENSG00000176946), score: 0.6 THUMPD1THUMP domain containing 1 (ENSG00000066654), score: -0.81 TIMM10translocase of inner mitochondrial membrane 10 homolog (yeast) (ENSG00000134809), score: 0.63 TIMM17Btranslocase of inner mitochondrial membrane 17 homolog B (yeast) (ENSG00000126768), score: 0.61 TMEM115transmembrane protein 115 (ENSG00000126062), score: 0.64 TMEM187transmembrane protein 187 (ENSG00000177854), score: 0.58 TMEM199transmembrane protein 199 (ENSG00000244045), score: -0.8 TMEM79transmembrane protein 79 (ENSG00000163472), score: 0.85 TMEM80transmembrane protein 80 (ENSG00000177042), score: 0.58 TMEM85transmembrane protein 85 (ENSG00000128463), score: 0.72 TRAP1TNF receptor-associated protein 1 (ENSG00000126602), score: 0.61 TRDMT1tRNA aspartic acid methyltransferase 1 (ENSG00000107614), score: -0.65 TRIM47tripartite motif-containing 47 (ENSG00000132481), score: 0.65 TRIM69tripartite motif-containing 69 (ENSG00000185880), score: 0.78 TRIP6thyroid hormone receptor interactor 6 (ENSG00000087077), score: 0.75 TRNT1tRNA nucleotidyl transferase, CCA-adding, 1 (ENSG00000072756), score: -0.78 TSG101tumor susceptibility gene 101 (ENSG00000074319), score: 0.57 TSNAXtranslin-associated factor X (ENSG00000116918), score: -0.64 TSPAN16tetraspanin 16 (ENSG00000130167), score: 0.82 TUBGCP2tubulin, gamma complex associated protein 2 (ENSG00000130640), score: 0.62 UPF2UPF2 regulator of nonsense transcripts homolog (yeast) (ENSG00000151461), score: -0.63 USP16ubiquitin specific peptidase 16 (ENSG00000156256), score: -0.63 UTP15UTP15, U3 small nucleolar ribonucleoprotein, homolog (S. cerevisiae) (ENSG00000164338), score: -0.61 UTS2urotensin 2 (ENSG00000049247), score: 0.8 WBP1WW domain binding protein 1 (ENSG00000115274), score: 0.59 WDR13WD repeat domain 13 (ENSG00000101940), score: 0.71 WDR18WD repeat domain 18 (ENSG00000065268), score: 0.58 WDR4WD repeat domain 4 (ENSG00000160193), score: 0.57 WDR67WD repeat domain 67 (ENSG00000156787), score: -0.64 WDR75WD repeat domain 75 (ENSG00000115368), score: -0.63 WDR83WD repeat domain 83 (ENSG00000123154), score: 0.67 WDR89WD repeat domain 89 (ENSG00000140006), score: -0.6 XPNPEP3X-prolyl aminopeptidase (aminopeptidase P) 3, putative (ENSG00000196236), score: 0.61 YIPF5Yip1 domain family, member 5 (ENSG00000145817), score: -0.67 YJEFN3YjeF N-terminal domain containing 3 (ENSG00000186010), score: 0.63 ZACNzinc activated ligand-gated ion channel (ENSG00000186919), score: 0.66 ZC3H6zinc finger CCCH-type containing 6 (ENSG00000188177), score: -0.77 ZFC3H1zinc finger, C3H1-type containing (ENSG00000133858), score: -0.62 ZMAT1zinc finger, matrin type 1 (ENSG00000166432), score: -0.74 ZNF200zinc finger protein 200 (ENSG00000010539), score: -0.63 ZNF214zinc finger protein 214 (ENSG00000149050), score: -0.61 ZNF23zinc finger protein 23 (KOX 16) (ENSG00000167377), score: -0.65 ZNF248zinc finger protein 248 (ENSG00000198105), score: -0.6 ZNF292zinc finger protein 292 (ENSG00000188994), score: -0.6 ZNF354Czinc finger protein 354C (ENSG00000177932), score: 0.7 ZNF444zinc finger protein 444 (ENSG00000167685), score: 0.58 ZNF518Bzinc finger protein 518B (ENSG00000178163), score: -0.61 ZNF557zinc finger protein 557 (ENSG00000130544), score: -0.66 ZNF581zinc finger protein 581 (ENSG00000171425), score: 0.73 ZNF599zinc finger protein 599 (ENSG00000153896), score: 0.67 ZNF76zinc finger protein 76 (expressed in testis) (ENSG00000065029), score: 0.6 ZNF776zinc finger protein 776 (ENSG00000152443), score: -0.61 ZUFSPzinc finger with UFM1-specific peptidase domain (ENSG00000153975), score: -0.64
| Id | species | tissue | sex | individual |
|---|---|---|---|---|
| ppa_br_m_ca1 | ppa | br | m | _ |
| hsa_cb_m_ca1 | hsa | cb | m | _ |
| hsa_cb_f_ca1 | hsa | cb | f | _ |
| hsa_ht_m2_ca1 | hsa | ht | m | 2 |
| hsa_ht_f_ca1 | hsa | ht | f | _ |
| hsa_lv_m1_ca1 | hsa | lv | m | 1 |
| hsa_br_m6_ca1 | hsa | br | m | 6 |
| hsa_kd_m2_ca1 | hsa | kd | m | 2 |
| hsa_br_m3_ca1 | hsa | br | m | 3 |
| hsa_kd_f_ca1 | hsa | kd | f | _ |
| hsa_lv_m2_ca1 | hsa | lv | m | 2 |
| hsa_ts_m1_ca1 | hsa | ts | m | 1 |
| hsa_ht_m1_ca1 | hsa | ht | m | 1 |
| hsa_br_m7_ca1 | hsa | br | m | 7 |
| hsa_kd_m1_ca1 | hsa | kd | m | 1 |