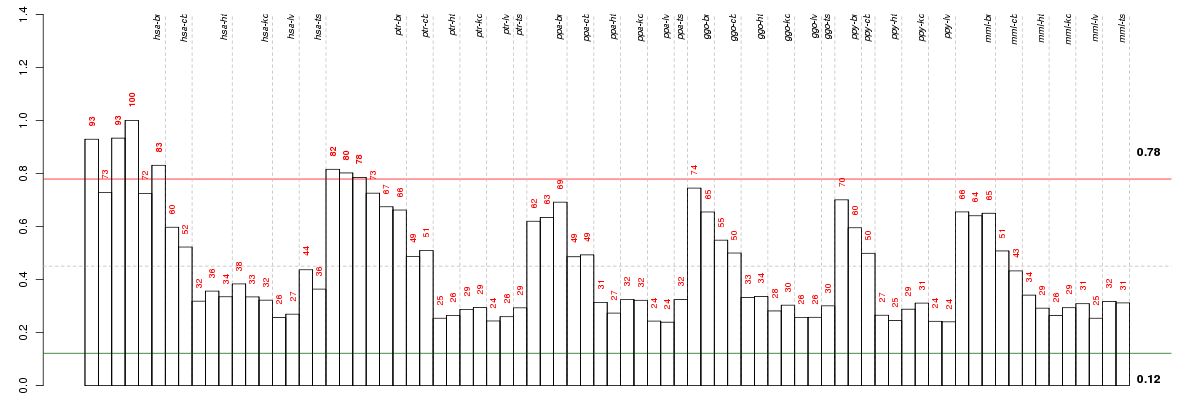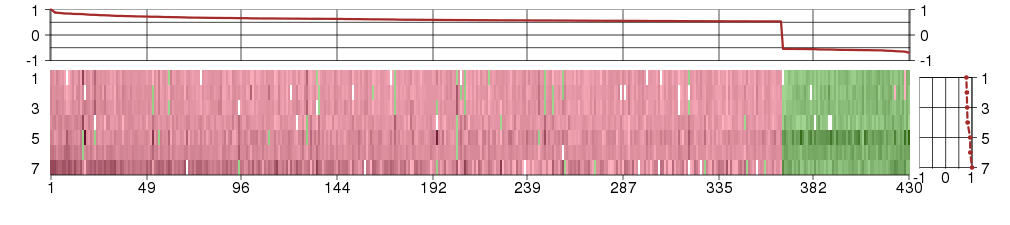

Under-expression is coded with green,
over-expression with red color.

cell morphogenesis
The developmental process by which the size or shape of a cell is generated and organized. Morphogenesis pertains to the creation of form.
cell morphogenesis involved in differentiation
The change in form (cell shape and size) that occurs when relatively unspecialized cells, e.g. embryonic or regenerative cells, acquire specialized structural and/or functional features that characterize the cells, tissues, or organs of the mature organism or some other relatively stable phase of the organism's life history.
regulation of action potential
Any process that modulates the frequency, rate or extent of action potential creation, propagation or termination. An action potential is a spike of membrane depolarization and repolarization that travels along the membrane of a cell.
system process
A multicellular organismal process carried out by any of the organs or tissues in an organ system. An organ system is a regularly interacting or interdependent group of organs or tissues that work together to carry out a biological objective.
signal transduction
The process whereby an activated receptor conveys information down the signaling pathway, resulting in a change in the function or state of a cell.
cellular ion homeostasis
Any process involved in the maintenance of an internal steady-state of ions at the level of a cell.
cell communication
Any process that mediates interactions between a cell and its surroundings. Encompasses interactions such as signaling or attachment between one cell and another cell, between a cell and an extracellular matrix, or between a cell and any other aspect of its environment.
small GTPase mediated signal transduction
Any series of molecular signals in which a small monomeric GTPase relays one or more of the signals.
cell-cell signaling
Any process that mediates the transfer of information from one cell to another.
synaptic transmission
The process of communication from a neuron to a target (neuron, muscle, or secretory cell) across a synapse.
multicellular organismal development
The biological process whose specific outcome is the progression of a multicellular organism over time from an initial condition (e.g. a zygote or a young adult) to a later condition (e.g. a multicellular animal or an aged adult).
anatomical structure morphogenesis
The process by which anatomical structures are generated and organized. Morphogenesis pertains to the creation of form.
nervous system development
The process whose specific outcome is the progression of nervous tissue over time, from its formation to its mature state.
neurological system process
A organ system process carried out by any of the organs or tissues of neurological system.
axonogenesis
Generation of a long process of a neuron, that carries efferent (outgoing) action potentials from the cell body towards target cells.
central nervous system development
The process whose specific outcome is the progression of the central nervous system over time, from its formation to the mature structure. The central nervous system is the core nervous system that serves an integrating and coordinating function. In vertebrates it consists of the brain, spinal cord and spinal nerves. In those invertebrates with a central nervous system it typically consists of a brain, cerebral ganglia and a nerve cord.
biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
cellular process
Any process that is carried out at the cellular level, but not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
glial cell differentiation
The process whereby a relatively unspecialized cell acquires the specialized features of a glial cell.
cellular component organization
A process that is carried out at the cellular level which results in the assembly, arrangement of constituent parts, or disassembly of a cellular component.
transmission of nerve impulse
The neurological system process by which a signal is transmitted through the nervous system by synaptic transmission and the sequential electrochemical polarization and depolarization that travels across the membrane of a nerve cell (neuron) in response to stimulation.
regulation of action potential in neuron
The process that modulates the membrane potential involved in the propagation of a signal in a neuron.
cellular homeostasis
Any process involved in the maintenance of an internal steady-state at the level of the cell.
neurogenesis
Generation of cells within the nervous system.
signaling pathway
The series of molecular events whereby information is sent from one location to another within a living organism or between living organisms.
intracellular signaling pathway
The series of molecular events whereby information is sent from one location to another within a cell.
signaling process
Any biological process involved in the generation, transmission, reception, or interpretation of a signal. A signal is an entity used to transmit or convey information.
signaling
The entirety of a process whereby information is transmitted. This process begins with the initiation of the signal and ends when a response has been triggered.
signal transmission
The process whereby a signal is released and/or conveyed from one location to another.
cell projection organization
A process that is carried out at the cellular level which results in the assembly, arrangement of constituent parts, or disassembly of a prolongation or process extending from a cell, e.g. a flagellum or axon.
cell differentiation
The process whereby relatively unspecialized cells, e.g. embryonic or regenerative cells, acquire specialized structural and/or functional features that characterize the cells, tissues, or organs of the mature organism or some other relatively stable phase of the organism's life history. Differentiation includes the processes involved in commitment of a cell to a specific fate and its subsequent development to the mature state.
neuron differentiation
The process whereby a relatively unspecialized cell acquires specialized features of a neuron.
neuron projection development
The process whose specific outcome is the progression of a neuron projection over time, from its formation to the mature structure. A neuron projection is any process extending from a neural cell, such as axons or dendrites (collectively called neurites).
multicellular organismal process
Any biological process, occurring at the level of a multicellular organism, pertinent to its function.
developmental process
A biological process whose specific outcome is the progression of an integrated living unit: an anatomical structure (which may be a subcellular structure, cell, tissue, or organ), or organism over time from an initial condition to a later condition.
cellular component morphogenesis
The process by which cellular structures, including whole cells or cell parts, are generated and organized. Morphogenesis pertains to the creation of form.
cell part morphogenesis
The process by which the anatomical structures of a cell part are generated and organized. Morphogenesis pertains to the creation of form.
intracellular signal transduction
The process whereby a signal is passed on to downstream components within the cell, which become activated themselves to further propagate the signal and finally trigger a change in the function or state of the cell.
gliogenesis
The process by which glial cells are generated. This includes the production of glial progenitors and their differentiation into mature glia.
regulation of membrane potential
Any process that modulates the establishment or extent of a membrane potential, the electric potential existing across any membrane arising from charges in the membrane itself and from the charges present in the media on either side of the membrane.
homeostatic process
Any biological process involved in the maintenance of an internal steady-state.
ion homeostasis
Any process involved in the maintenance of an internal steady-state of ions within an organism or cell.
cell development
The process whose specific outcome is the progression of the cell over time, from its formation to the mature structure. Cell development does not include the steps involved in committing a cell to a specific fate.
neuron development
The process whose specific outcome is the progression of a neuron over time, from initial commitment of the cell to a specific fate, to the fully functional differentiated cell.
cell morphogenesis involved in neuron differentiation
The process by which the structures of a neuron are generated and organized. This process occurs while the initially relatively unspecialized cell is acquiring the specialized features of a neuron.
generation of neurons
The process by which nerve cells are generated. This includes the production of neuroblasts and their differentiation into neurons.
oligodendrocyte differentiation
The process whereby a relatively unspecialized cell acquires the specialized features of an oligodendrocyte. An oligodendrocyte is a type of glial cell involved in myelinating the axons of neurons in the central nervous system.
system development
The process whose specific outcome is the progression of an organismal system over time, from its formation to the mature structure. A system is a regularly interacting or interdependent group of organs or tissues that work together to carry out a given biological process.
neuron projection morphogenesis
The process by which the anatomical structures of a neuron projection are generated and organized. Morphogenesis pertains to the creation of form. A neuron projection is any process extending from a neural cell, such as axons or dendrites.
anatomical structure development
The biological process whose specific outcome is the progression of an anatomical structure from an initial condition to its mature state. This process begins with the formation of the structure and ends with the mature structure, whatever form that may be including its natural destruction. An anatomical structure is any biological entity that occupies space and is distinguished from its surroundings. Anatomical structures can be macroscopic such as a carpel, or microscopic such as an acrosome.
cell projection morphogenesis
The process by which the anatomical structures of a cell projection are generated and organized. Morphogenesis pertains to the creation of form.
cellular developmental process
A biological process whose specific outcome is the progression of a cell over time from an initial condition to a later condition.
chemical homeostasis
Any biological process involved in the maintenance of an internal steady-state of a chemical.
regulation of biological process
Any process that modulates the frequency, rate or extent of a biological process. Biological processes are regulated by many means; examples include the control of gene expression, protein modification or interaction with a protein or substrate molecule.
regulation of cellular process
Any process that modulates the frequency, rate or extent of a cellular process, any of those that are carried out at the cellular level, but are not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
cellular chemical homeostasis
Any biological process involved in the maintenance of an internal steady-state of a chemical at the level of the cell.
biological regulation
Any process that modulates the frequency, rate or extent of any biological process, quality or function.
regulation of biological quality
Any process that modulates the frequency, rate or extent of a biological quality. A biological quality is a measurable attribute of an organism or part of an organism, such as size, mass, shape, color, etc.
all
NA
cell projection organization
A process that is carried out at the cellular level which results in the assembly, arrangement of constituent parts, or disassembly of a prolongation or process extending from a cell, e.g. a flagellum or axon.
signaling process
Any biological process involved in the generation, transmission, reception, or interpretation of a signal. A signal is an entity used to transmit or convey information.
multicellular organismal development
The biological process whose specific outcome is the progression of a multicellular organism over time from an initial condition (e.g. a zygote or a young adult) to a later condition (e.g. a multicellular animal or an aged adult).
cellular developmental process
A biological process whose specific outcome is the progression of a cell over time from an initial condition to a later condition.
regulation of cellular process
Any process that modulates the frequency, rate or extent of a cellular process, any of those that are carried out at the cellular level, but are not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
regulation of biological process
Any process that modulates the frequency, rate or extent of a biological process. Biological processes are regulated by many means; examples include the control of gene expression, protein modification or interaction with a protein or substrate molecule.
cell-cell signaling
Any process that mediates the transfer of information from one cell to another.
cellular component morphogenesis
The process by which cellular structures, including whole cells or cell parts, are generated and organized. Morphogenesis pertains to the creation of form.
signal transduction
The process whereby an activated receptor conveys information down the signaling pathway, resulting in a change in the function or state of a cell.
transmission of nerve impulse
The neurological system process by which a signal is transmitted through the nervous system by synaptic transmission and the sequential electrochemical polarization and depolarization that travels across the membrane of a nerve cell (neuron) in response to stimulation.
cellular component morphogenesis
The process by which cellular structures, including whole cells or cell parts, are generated and organized. Morphogenesis pertains to the creation of form.
anatomical structure morphogenesis
The process by which anatomical structures are generated and organized. Morphogenesis pertains to the creation of form.
cell development
The process whose specific outcome is the progression of the cell over time, from its formation to the mature structure. Cell development does not include the steps involved in committing a cell to a specific fate.
system development
The process whose specific outcome is the progression of an organismal system over time, from its formation to the mature structure. A system is a regularly interacting or interdependent group of organs or tissues that work together to carry out a given biological process.
synaptic transmission
The process of communication from a neuron to a target (neuron, muscle, or secretory cell) across a synapse.
neuron projection morphogenesis
The process by which the anatomical structures of a neuron projection are generated and organized. Morphogenesis pertains to the creation of form. A neuron projection is any process extending from a neural cell, such as axons or dendrites.
cell development
The process whose specific outcome is the progression of the cell over time, from its formation to the mature structure. Cell development does not include the steps involved in committing a cell to a specific fate.
cell morphogenesis involved in differentiation
The change in form (cell shape and size) that occurs when relatively unspecialized cells, e.g. embryonic or regenerative cells, acquire specialized structural and/or functional features that characterize the cells, tissues, or organs of the mature organism or some other relatively stable phase of the organism's life history.
cell projection morphogenesis
The process by which the anatomical structures of a cell projection are generated and organized. Morphogenesis pertains to the creation of form.
cell projection morphogenesis
The process by which the anatomical structures of a cell projection are generated and organized. Morphogenesis pertains to the creation of form.
intracellular signal transduction
The process whereby a signal is passed on to downstream components within the cell, which become activated themselves to further propagate the signal and finally trigger a change in the function or state of the cell.
transmission of nerve impulse
The neurological system process by which a signal is transmitted through the nervous system by synaptic transmission and the sequential electrochemical polarization and depolarization that travels across the membrane of a nerve cell (neuron) in response to stimulation.
cellular homeostasis
Any process involved in the maintenance of an internal steady-state at the level of the cell.
neuron development
The process whose specific outcome is the progression of a neuron over time, from initial commitment of the cell to a specific fate, to the fully functional differentiated cell.
neuron projection development
The process whose specific outcome is the progression of a neuron projection over time, from its formation to the mature structure. A neuron projection is any process extending from a neural cell, such as axons or dendrites (collectively called neurites).
cell morphogenesis involved in neuron differentiation
The process by which the structures of a neuron are generated and organized. This process occurs while the initially relatively unspecialized cell is acquiring the specialized features of a neuron.
central nervous system development
The process whose specific outcome is the progression of the central nervous system over time, from its formation to the mature structure. The central nervous system is the core nervous system that serves an integrating and coordinating function. In vertebrates it consists of the brain, spinal cord and spinal nerves. In those invertebrates with a central nervous system it typically consists of a brain, cerebral ganglia and a nerve cord.
neurogenesis
Generation of cells within the nervous system.
oligodendrocyte differentiation
The process whereby a relatively unspecialized cell acquires the specialized features of an oligodendrocyte. An oligodendrocyte is a type of glial cell involved in myelinating the axons of neurons in the central nervous system.
cellular chemical homeostasis
Any biological process involved in the maintenance of an internal steady-state of a chemical at the level of the cell.
glial cell differentiation
The process whereby a relatively unspecialized cell acquires the specialized features of a glial cell.
neuron differentiation
The process whereby a relatively unspecialized cell acquires specialized features of a neuron.
axonogenesis
Generation of a long process of a neuron, that carries efferent (outgoing) action potentials from the cell body towards target cells.
cellular ion homeostasis
Any process involved in the maintenance of an internal steady-state of ions at the level of a cell.
regulation of action potential in neuron
The process that modulates the membrane potential involved in the propagation of a signal in a neuron.

intracellular
The living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
membrane
Double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
integral to membrane
Penetrating at least one phospholipid bilayer of a membrane. May also refer to the state of being buried in the bilayer with no exposure outside the bilayer. When used to describe a protein, indicates that all or part of the peptide sequence is embedded in the membrane.
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
cell
The basic structural and functional unit of all organisms. Includes the plasma membrane and any external encapsulating structures such as the cell wall and cell envelope.
cytoplasm
All of the contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures.
synaptic vesicle
A secretory organelle, some 50 nm in diameter, of presynaptic nerve terminals; accumulates in high concentrations of neurotransmitters and is secreted these into the synaptic cleft by fusion with the 'active zone' of the presynaptic plasma membrane.
cytoplasmic membrane-bounded vesicle
A membrane-bounded vesicle found in the cytoplasm of the cell.
coated vesicle
Small membrane-bounded organelle formed by pinching off of a coated region of membrane. Some coats are made of clathrin, whereas others are made from other proteins.
clathrin-coated vesicle
A vesicle with a coat formed of clathrin connected to the membrane via one of the clathrin adaptor complexes.
axon
The long process of a neuron that conducts nerve impulses, usually away from the cell body to the terminals and varicosities, which are sites of storage and release of neurotransmitter.
intrinsic to membrane
Located in a membrane such that some covalently attached portion of the gene product, for example part of a peptide sequence or some other covalently attached moiety such as a GPI anchor, spans or is embedded in one or both leaflets of the membrane.
cytoplasmic vesicle
A vesicle formed of membrane or protein, found in the cytoplasm of a cell.
vesicle
Any small, fluid-filled, spherical organelle enclosed by membrane or protein.
membrane-bounded vesicle
Any small, fluid-filled, spherical organelle enclosed by a lipid bilayer.
axon part
A part of an axon, a cell projection of a neuron.
cell projection
A prolongation or process extending from a cell, e.g. a flagellum or axon.
neuron projection
A prolongation or process extending from a nerve cell, e.g. an axon or dendrite.
myelin sheath
An electrically insulating fatty layer that surrounds the axons of many neurons. It is an outgrowth of glial cells: Schwann cells supply the myelin for peripheral neurons while oligodendrocytes supply it to those of the central nervous system.
compact myelin
The portion of the myelin sheath in which layers of cell membrane are tightly juxtaposed, completely excluding cytoplasm. The juxtaposed cytoplasmic surfaces form the major dense line, while the juxtaposed extracellular surfaces form the interperiod line visible in electron micrographs.
organelle
Organized structure of distinctive morphology and function. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton, and prokaryotic structures such as anammoxosomes and pirellulosomes. Excludes the plasma membrane.
membrane-bounded organelle
Organized structure of distinctive morphology and function, bounded by a single or double lipid bilayer membrane. Includes the nucleus, mitochondria, plastids, vacuoles, and vesicles. Excludes the plasma membrane.
intracellular organelle
Organized structure of distinctive morphology and function, occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton. Excludes the plasma membrane.
intracellular membrane-bounded organelle
Organized structure of distinctive morphology and function, bounded by a single or double lipid bilayer membrane and occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, and vesicles. Excludes the plasma membrane.
axon terminus
Terminal inflated portion of the axon, containing the specialized apparatus necessary to release neurotransmitters. The axon terminus is considered to be the whole region of thickening and the terminal button is a specialized region of it.
neuron projection terminus
The specialized, terminal region of a neuron projection such as an axon or a dendrite.
intracellular part
Any constituent part of the living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
membrane part
Any constituent part of a membrane, a double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
cytoplasmic part
Any constituent part of the cytoplasm, all of the contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures.
synapse part
Any constituent part of a synapse, the junction between a nerve fiber of one neuron and another neuron or muscle fiber or glial cell.
cell projection part
Any constituent part of a cell projection, a prolongation or process extending from a cell, e.g. a flagellum or axon.
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
synapse
The junction between a nerve fiber of one neuron and another neuron or muscle fiber or glial cell; the site of interneuronal communication. As the nerve fiber approaches the synapse it enlarges into a specialized structure, the presynaptic nerve ending, which contains mitochondria and synaptic vesicles. At the tip of the nerve ending is the presynaptic membrane; facing it, and separated from it by a minute cleft (the synaptic cleft) is a specialized area of membrane on the receiving cell, known as the postsynaptic membrane. In response to the arrival of nerve impulses, the presynaptic nerve ending secretes molecules of neurotransmitters into the synaptic cleft. These diffuse across the cleft and transmit the signal to the postsynaptic membrane.
all
NA
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
synapse part
Any constituent part of a synapse, the junction between a nerve fiber of one neuron and another neuron or muscle fiber or glial cell.
cytoplasmic vesicle
A vesicle formed of membrane or protein, found in the cytoplasm of a cell.
intracellular membrane-bounded organelle
Organized structure of distinctive morphology and function, bounded by a single or double lipid bilayer membrane and occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, and vesicles. Excludes the plasma membrane.
intracellular part
Any constituent part of the living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
membrane part
Any constituent part of a membrane, a double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
cell projection part
Any constituent part of a cell projection, a prolongation or process extending from a cell, e.g. a flagellum or axon.
compact myelin
The portion of the myelin sheath in which layers of cell membrane are tightly juxtaposed, completely excluding cytoplasm. The juxtaposed cytoplasmic surfaces form the major dense line, while the juxtaposed extracellular surfaces form the interperiod line visible in electron micrographs.
intracellular organelle
Organized structure of distinctive morphology and function, occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton. Excludes the plasma membrane.
cytoplasmic membrane-bounded vesicle
A membrane-bounded vesicle found in the cytoplasm of the cell.
cytoplasmic membrane-bounded vesicle
A membrane-bounded vesicle found in the cytoplasm of the cell.
neuron projection terminus
The specialized, terminal region of a neuron projection such as an axon or a dendrite.
cytoplasmic part
Any constituent part of the cytoplasm, all of the contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures.
cytoplasmic vesicle
A vesicle formed of membrane or protein, found in the cytoplasm of a cell.
axon terminus
Terminal inflated portion of the axon, containing the specialized apparatus necessary to release neurotransmitters. The axon terminus is considered to be the whole region of thickening and the terminal button is a specialized region of it.
axon part
A part of an axon, a cell projection of a neuron.
synaptic vesicle
A secretory organelle, some 50 nm in diameter, of presynaptic nerve terminals; accumulates in high concentrations of neurotransmitters and is secreted these into the synaptic cleft by fusion with the 'active zone' of the presynaptic plasma membrane.

AATKapoptosis-associated tyrosine kinase (ENSG00000181409), score: 0.6 ABCA2ATP-binding cassette, sub-family A (ABC1), member 2 (ENSG00000107331), score: 0.73 ABHD12abhydrolase domain containing 12 (ENSG00000100997), score: 0.58 ACTR1AARP1 actin-related protein 1 homolog A, centractin alpha (yeast) (ENSG00000138107), score: 0.57 ADAMTS14ADAM metallopeptidase with thrombospondin type 1 motif, 14 (ENSG00000138316), score: 0.75 ADAMTS4ADAM metallopeptidase with thrombospondin type 1 motif, 4 (ENSG00000158859), score: 0.61 ADAMTS8ADAM metallopeptidase with thrombospondin type 1 motif, 8 (ENSG00000134917), score: 0.63 ADCK1aarF domain containing kinase 1 (ENSG00000063761), score: 0.64 ADORA1adenosine A1 receptor (ENSG00000163485), score: 0.55 AGAP1ArfGAP with GTPase domain, ankyrin repeat and PH domain 1 (ENSG00000157985), score: 0.57 AGAP3ArfGAP with GTPase domain, ankyrin repeat and PH domain 3 (ENSG00000133612), score: 0.54 AGPAT41-acylglycerol-3-phosphate O-acyltransferase 4 (lysophosphatidic acid acyltransferase, delta) (ENSG00000026652), score: 0.65 AK5adenylate kinase 5 (ENSG00000154027), score: 0.59 ANLNanillin, actin binding protein (ENSG00000011426), score: 0.68 ANO4anoctamin 4 (ENSG00000151572), score: 0.61 AP3M1adaptor-related protein complex 3, mu 1 subunit (ENSG00000185009), score: -0.55 APLP1amyloid beta (A4) precursor-like protein 1 (ENSG00000105290), score: 0.59 ARCactivity-regulated cytoskeleton-associated protein (ENSG00000198576), score: 0.54 ARHGAP22Rho GTPase activating protein 22 (ENSG00000128805), score: 0.67 ARHGAP29Rho GTPase activating protein 29 (ENSG00000137962), score: -0.65 ARHGAP36Rho GTPase activating protein 36 (ENSG00000147256), score: 0.6 ARHGEF2Rho/Rac guanine nucleotide exchange factor (GEF) 2 (ENSG00000116584), score: 0.55 ARL8AADP-ribosylation factor-like 8A (ENSG00000143862), score: 0.57 ARRDC2arrestin domain containing 2 (ENSG00000105643), score: 0.6 ASPRV1aspartic peptidase, retroviral-like 1 (ENSG00000244617), score: 0.55 ATL2atlastin GTPase 2 (ENSG00000119787), score: -0.55 ATP10BATPase, class V, type 10B (ENSG00000118322), score: 0.78 ATP6V0D1ATPase, H+ transporting, lysosomal 38kDa, V0 subunit d1 (ENSG00000159720), score: 0.56 B3GAT1beta-1,3-glucuronyltransferase 1 (glucuronosyltransferase P) (ENSG00000109956), score: 0.57 B4GALT2UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 2 (ENSG00000117411), score: 0.58 BACE1beta-site APP-cleaving enzyme 1 (ENSG00000186318), score: 0.63 BAI2brain-specific angiogenesis inhibitor 2 (ENSG00000121753), score: 0.53 BCANbrevican (ENSG00000132692), score: 0.54 BCAS1breast carcinoma amplified sequence 1 (ENSG00000064787), score: 0.63 BCCIPBRCA2 and CDKN1A interacting protein (ENSG00000107949), score: -0.58 BIN1bridging integrator 1 (ENSG00000136717), score: 0.57 C10orf90chromosome 10 open reading frame 90 (ENSG00000154493), score: 0.58 C11orf9chromosome 11 open reading frame 9 (ENSG00000124920), score: 0.79 C12orf76chromosome 12 open reading frame 76 (ENSG00000174456), score: 0.58 C16orf13chromosome 16 open reading frame 13 (ENSG00000130731), score: 0.54 C16orf70chromosome 16 open reading frame 70 (ENSG00000125149), score: 0.53 C19orf62chromosome 19 open reading frame 62 (ENSG00000105393), score: 0.53 C1orf128chromosome 1 open reading frame 128 (ENSG00000057757), score: 0.56 C1orf198chromosome 1 open reading frame 198 (ENSG00000119280), score: 0.75 C1orf95chromosome 1 open reading frame 95 (ENSG00000203685), score: 0.55 C1QL2complement component 1, q subcomponent-like 2 (ENSG00000144119), score: 0.55 C20orf103chromosome 20 open reading frame 103 (ENSG00000125869), score: 0.55 C22orf9chromosome 22 open reading frame 9 (ENSG00000100364), score: 0.75 C2orf82chromosome 2 open reading frame 82 (ENSG00000182600), score: 0.63 C5orf30chromosome 5 open reading frame 30 (ENSG00000181751), score: 0.58 C6orf1chromosome 6 open reading frame 1 (ENSG00000186577), score: 0.63 C7orf41chromosome 7 open reading frame 41 (ENSG00000180354), score: 0.59 C8orf46chromosome 8 open reading frame 46 (ENSG00000169085), score: 0.55 CALHM1calcium homeostasis modulator 1 (ENSG00000185933), score: 0.66 CALYcalcyon neuron-specific vesicular protein (ENSG00000130643), score: 0.54 CAPN3calpain 3, (p94) (ENSG00000092529), score: 0.73 CAPN9calpain 9 (ENSG00000135773), score: 0.78 CCT8chaperonin containing TCP1, subunit 8 (theta) (ENSG00000156261), score: -0.65 CD22CD22 molecule (ENSG00000012124), score: 0.95 CD2APCD2-associated protein (ENSG00000198087), score: -0.6 CDC23cell division cycle 23 homolog (S. cerevisiae) (ENSG00000094880), score: -0.58 CDC26cell division cycle 26 homolog (S. cerevisiae) (ENSG00000176386), score: -0.55 CDC42EP2CDC42 effector protein (Rho GTPase binding) 2 (ENSG00000149798), score: 0.56 CDH20cadherin 20, type 2 (ENSG00000101542), score: 0.6 CDH4cadherin 4, type 1, R-cadherin (retinal) (ENSG00000179242), score: 0.57 CDHR1cadherin-related family member 1 (ENSG00000148600), score: 0.66 CDK18cyclin-dependent kinase 18 (ENSG00000117266), score: 0.55 CDK2cyclin-dependent kinase 2 (ENSG00000123374), score: -0.67 CDK2AP1cyclin-dependent kinase 2 associated protein 1 (ENSG00000111328), score: 0.62 CELSR2cadherin, EGF LAG seven-pass G-type receptor 2 (flamingo homolog, Drosophila) (ENSG00000143126), score: 0.53 CERCAMcerebral endothelial cell adhesion molecule (ENSG00000167123), score: 0.82 CHADLchondroadherin-like (ENSG00000100399), score: 0.79 CHD1chromodomain helicase DNA binding protein 1 (ENSG00000153922), score: -0.57 CHGAchromogranin A (parathyroid secretory protein 1) (ENSG00000100604), score: 0.57 CHRM1cholinergic receptor, muscarinic 1 (ENSG00000168539), score: 0.58 CHST6carbohydrate (N-acetylglucosamine 6-O) sulfotransferase 6 (ENSG00000183196), score: 0.64 CLCA4chloride channel accessory 4 (ENSG00000016602), score: 0.88 CLDN11claudin 11 (ENSG00000013297), score: 0.68 CLDND1claudin domain containing 1 (ENSG00000080822), score: 0.77 CLN8ceroid-lipofuscinosis, neuronal 8 (epilepsy, progressive with mental retardation) (ENSG00000182372), score: 0.7 CLUL1clusterin-like 1 (retinal) (ENSG00000079101), score: 0.61 CMTM5CKLF-like MARVEL transmembrane domain containing 5 (ENSG00000166091), score: 0.74 CNDP1carnosine dipeptidase 1 (metallopeptidase M20 family) (ENSG00000150656), score: 0.54 CNP2',3'-cyclic nucleotide 3' phosphodiesterase (ENSG00000173786), score: 0.86 CNTN2contactin 2 (axonal) (ENSG00000184144), score: 0.62 COBLcordon-bleu homolog (mouse) (ENSG00000106078), score: 0.58 COL11A2collagen, type XI, alpha 2 (ENSG00000204248), score: 0.55 COL16A1collagen, type XVI, alpha 1 (ENSG00000084636), score: 0.63 COL20A1collagen, type XX, alpha 1 (ENSG00000101203), score: 0.64 COL9A2collagen, type IX, alpha 2 (ENSG00000049089), score: 0.65 COPS7BCOP9 constitutive photomorphogenic homolog subunit 7B (Arabidopsis) (ENSG00000144524), score: 0.6 CPNE2copine II (ENSG00000140848), score: 0.72 CPNE6copine VI (neuronal) (ENSG00000100884), score: 0.53 CREG2cellular repressor of E1A-stimulated genes 2 (ENSG00000175874), score: 0.55 CRYBB1crystallin, beta B1 (ENSG00000100122), score: 0.59 CSRP1cysteine and glycine-rich protein 1 (ENSG00000159176), score: 0.61 CTNNAL1catenin (cadherin-associated protein), alpha-like 1 (ENSG00000119326), score: -0.55 CTR9Ctr9, Paf1/RNA polymerase II complex component, homolog (S. cerevisiae) (ENSG00000198730), score: -0.59 CWC22CWC22 spliceosome-associated protein homolog (S. cerevisiae) (ENSG00000163510), score: -0.57 CYB5D1cytochrome b5 domain containing 1 (ENSG00000182224), score: -0.63 CYP46A1cytochrome P450, family 46, subfamily A, polypeptide 1 (ENSG00000036530), score: 0.54 DAAM2dishevelled associated activator of morphogenesis 2 (ENSG00000146122), score: 0.79 DBNDD2dysbindin (dystrobrevin binding protein 1) domain containing 2 (ENSG00000244274), score: 0.56 DDX3XDEAD (Asp-Glu-Ala-Asp) box polypeptide 3, X-linked (ENSG00000215301), score: -0.6 DEFB134defensin, beta 134 (ENSG00000205882), score: 0.57 DMBT1deleted in malignant brain tumors 1 (ENSG00000187908), score: 0.63 DNAJB2DnaJ (Hsp40) homolog, subfamily B, member 2 (ENSG00000135924), score: 0.54 DNAJC7DnaJ (Hsp40) homolog, subfamily C, member 7 (ENSG00000168259), score: 0.57 DNMBPdynamin binding protein (ENSG00000107554), score: -0.64 DOHHdeoxyhypusine hydroxylase/monooxygenase (ENSG00000129932), score: 0.68 DPYSL5dihydropyrimidinase-like 5 (ENSG00000157851), score: 0.54 DSCAML1Down syndrome cell adhesion molecule like 1 (ENSG00000177103), score: 0.62 DTNBdystrobrevin, beta (ENSG00000138101), score: 0.57 DUOXA1dual oxidase maturation factor 1 (ENSG00000140254), score: 0.64 E2F1E2F transcription factor 1 (ENSG00000101412), score: 0.59 EDIL3EGF-like repeats and discoidin I-like domains 3 (ENSG00000164176), score: 0.64 EDN3endothelin 3 (ENSG00000124205), score: 0.57 EFSembryonal Fyn-associated substrate (ENSG00000100842), score: 0.69 EIF3Eeukaryotic translation initiation factor 3, subunit E (ENSG00000104408), score: -0.57 EIF4EBP2eukaryotic translation initiation factor 4E binding protein 2 (ENSG00000148730), score: -0.64 ELOVL1elongation of very long chain fatty acids (FEN1/Elo2, SUR4/Elo3, yeast)-like 1 (ENSG00000066322), score: 0.76 EML2echinoderm microtubule associated protein like 2 (ENSG00000125746), score: 0.73 ENDOD1endonuclease domain containing 1 (ENSG00000149218), score: 0.66 ENPP2ectonucleotide pyrophosphatase/phosphodiesterase 2 (ENSG00000136960), score: 0.73 EPN2epsin 2 (ENSG00000072134), score: 0.54 ERMNermin, ERM-like protein (ENSG00000136541), score: 0.7 EVI2Aecotropic viral integration site 2A (ENSG00000126860), score: 0.8 FA2Hfatty acid 2-hydroxylase (ENSG00000103089), score: 0.83 FAM102Afamily with sequence similarity 102, member A (ENSG00000167106), score: 0.68 FAM108B1family with sequence similarity 108, member B1 (ENSG00000107362), score: 0.68 FAM114A2family with sequence similarity 114, member A2 (ENSG00000055147), score: -0.55 FAM124Afamily with sequence similarity 124A (ENSG00000150510), score: 0.84 FAM125Bfamily with sequence similarity 125, member B (ENSG00000196814), score: 0.64 FAM13Cfamily with sequence similarity 13, member C (ENSG00000148541), score: 0.59 FAM171A1family with sequence similarity 171, member A1 (ENSG00000148468), score: 0.53 FAM53Bfamily with sequence similarity 53, member B (ENSG00000189319), score: 0.54 FAM5Bfamily with sequence similarity 5, member B (ENSG00000198797), score: 0.54 FCHO1FCH domain only 1 (ENSG00000130475), score: 0.63 FEZ1fasciculation and elongation protein zeta 1 (zygin I) (ENSG00000149557), score: 0.57 FEZF2FEZ family zinc finger 2 (ENSG00000153266), score: 0.62 FFAR1free fatty acid receptor 1 (ENSG00000126266), score: 0.79 FGF22fibroblast growth factor 22 (ENSG00000070388), score: 0.7 FKBP8FK506 binding protein 8, 38kDa (ENSG00000105701), score: 0.57 FLRT1fibronectin leucine rich transmembrane protein 1 (ENSG00000126500), score: 0.55 FNBP1Lformin binding protein 1-like (ENSG00000137942), score: -0.64 FREM3FRAS1 related extracellular matrix 3 (ENSG00000183090), score: 0.54 FSCN1fascin homolog 1, actin-bundling protein (Strongylocentrotus purpuratus) (ENSG00000075618), score: 0.67 FZD6frizzled homolog 6 (Drosophila) (ENSG00000164930), score: -0.55 GAB2GRB2-associated binding protein 2 (ENSG00000033327), score: 0.71 GABRQgamma-aminobutyric acid (GABA) receptor, theta (ENSG00000147402), score: 0.54 GAL3ST1galactose-3-O-sulfotransferase 1 (ENSG00000128242), score: 0.65 GALNT6UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 6 (GalNAc-T6) (ENSG00000139629), score: 0.84 GIPC1GIPC PDZ domain containing family, member 1 (ENSG00000123159), score: 0.65 GJD4gap junction protein, delta 4, 40.1kDa (ENSG00000177291), score: 0.68 GLDNgliomedin (ENSG00000186417), score: 0.82 GLT25D2glycosyltransferase 25 domain containing 2 (ENSG00000198756), score: 0.6 GLTPglycolipid transfer protein (ENSG00000139433), score: 0.61 GNAI1guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 1 (ENSG00000127955), score: 0.57 GOLGA7golgin A7 (ENSG00000147533), score: 0.65 GPM6Bglycoprotein M6B (ENSG00000046653), score: 0.55 GPR123G protein-coupled receptor 123 (ENSG00000197177), score: 0.62 GPR151G protein-coupled receptor 151 (ENSG00000173250), score: 0.59 GPR37G protein-coupled receptor 37 (endothelin receptor type B-like) (ENSG00000170775), score: 0.72 GPR78G protein-coupled receptor 78 (ENSG00000155269), score: 0.81 GPRC5BG protein-coupled receptor, family C, group 5, member B (ENSG00000167191), score: 0.58 GREM1gremlin 1 (ENSG00000166923), score: 0.59 GRID1glutamate receptor, ionotropic, delta 1 (ENSG00000182771), score: 0.63 GRIK4glutamate receptor, ionotropic, kainate 4 (ENSG00000149403), score: 0.54 GRM3glutamate receptor, metabotropic 3 (ENSG00000198822), score: 0.62 GSPT1G1 to S phase transition 1 (ENSG00000103342), score: -0.62 GUK1guanylate kinase 1 (ENSG00000143774), score: 0.64 HDAC7histone deacetylase 7 (ENSG00000061273), score: 0.54 HELQhelicase, POLQ-like (ENSG00000163312), score: -0.55 HHIPhedgehog interacting protein (ENSG00000164161), score: 0.54 HHIPL1HHIP-like 1 (ENSG00000182218), score: 0.54 HIPK2homeodomain interacting protein kinase 2 (ENSG00000064393), score: 0.56 HS3ST2heparan sulfate (glucosamine) 3-O-sulfotransferase 2 (ENSG00000122254), score: 0.57 HS3ST4heparan sulfate (glucosamine) 3-O-sulfotransferase 4 (ENSG00000182601), score: 0.59 HSD11B1Lhydroxysteroid (11-beta) dehydrogenase 1-like (ENSG00000167733), score: 0.57 HTRA1HtrA serine peptidase 1 (ENSG00000166033), score: 0.55 ICOSLGinducible T-cell co-stimulator ligand (ENSG00000160223), score: 0.67 IDEinsulin-degrading enzyme (ENSG00000119912), score: -0.6 IFNA2interferon, alpha 2 (ENSG00000188379), score: 0.59 IGSF8immunoglobulin superfamily, member 8 (ENSG00000162729), score: 0.56 IL1RAPL1interleukin 1 receptor accessory protein-like 1 (ENSG00000169306), score: 0.61 IPO13importin 13 (ENSG00000117408), score: 0.54 ITPK1inositol 1,3,4-triphosphate 5/6 kinase (ENSG00000100605), score: 0.61 JAK1Janus kinase 1 (ENSG00000162434), score: -0.62 JAM3junctional adhesion molecule 3 (ENSG00000166086), score: 0.61 KCNF1potassium voltage-gated channel, subfamily F, member 1 (ENSG00000162975), score: 0.58 KCNH3potassium voltage-gated channel, subfamily H (eag-related), member 3 (ENSG00000135519), score: 0.55 KCNH8potassium voltage-gated channel, subfamily H (eag-related), member 8 (ENSG00000183960), score: 0.88 KCNMB4potassium large conductance calcium-activated channel, subfamily M, beta member 4 (ENSG00000135643), score: 0.56 KCNN1potassium intermediate/small conductance calcium-activated channel, subfamily N, member 1 (ENSG00000105642), score: 0.56 KCTD4potassium channel tetramerisation domain containing 4 (ENSG00000180332), score: 0.59 KIAA1429KIAA1429 (ENSG00000164944), score: -0.59 KIAA1598KIAA1598 (ENSG00000187164), score: 0.56 KINKIN, antigenic determinant of recA protein homolog (mouse) (ENSG00000151657), score: -0.6 KIRREL3kin of IRRE like 3 (Drosophila) (ENSG00000149571), score: 0.53 KLF11Kruppel-like factor 11 (ENSG00000172059), score: -0.62 KLHDC5kelch domain containing 5 (ENSG00000087448), score: 0.53 KLHL26kelch-like 26 (Drosophila) (ENSG00000167487), score: 0.58 KLHL32kelch-like 32 (Drosophila) (ENSG00000186231), score: 0.6 KLHL4kelch-like 4 (Drosophila) (ENSG00000102271), score: 0.58 KREMEN2kringle containing transmembrane protein 2 (ENSG00000131650), score: 0.84 KRT83keratin 83 (ENSG00000170523), score: 0.65 L3MBTL2l(3)mbt-like 2 (Drosophila) (ENSG00000100395), score: 0.56 LANCL1LanC lantibiotic synthetase component C-like 1 (bacterial) (ENSG00000115365), score: 0.58 LAPTM4Alysosomal protein transmembrane 4 alpha (ENSG00000068697), score: -0.59 LAYNlayilin (ENSG00000204381), score: 0.59 LGI3leucine-rich repeat LGI family, member 3 (ENSG00000168481), score: 0.59 LGR5leucine-rich repeat-containing G protein-coupled receptor 5 (ENSG00000139292), score: 0.83 LHPPphospholysine phosphohistidine inorganic pyrophosphate phosphatase (ENSG00000107902), score: 0.61 LIMK1LIM domain kinase 1 (ENSG00000106683), score: 0.53 LIX1Lix1 homolog (chicken) (ENSG00000145721), score: 0.63 LNX2ligand of numb-protein X 2 (ENSG00000139517), score: -0.59 LOC100294412similar to KIAA0655 protein (ENSG00000130787), score: 0.63 LPPR3lipid phosphate phosphatase-related protein type 3 (ENSG00000129951), score: 0.62 LRIT2leucine-rich repeat, immunoglobulin-like and transmembrane domains 2 (ENSG00000204033), score: 0.72 LY6Hlymphocyte antigen 6 complex, locus H (ENSG00000176956), score: 0.57 LYPD5LY6/PLAUR domain containing 5 (ENSG00000159871), score: 0.65 LZTS2leucine zipper, putative tumor suppressor 2 (ENSG00000107816), score: 0.55 MAGmyelin associated glycoprotein (ENSG00000105695), score: 0.74 MALT1mucosa associated lymphoid tissue lymphoma translocation gene 1 (ENSG00000172175), score: -0.59 MAP3K1mitogen-activated protein kinase kinase kinase 1 (ENSG00000095015), score: -0.55 MAP4K4mitogen-activated protein kinase kinase kinase kinase 4 (ENSG00000071054), score: 0.56 MAPK3mitogen-activated protein kinase 3 (ENSG00000102882), score: 0.57 MARCH1membrane-associated ring finger (C3HC4) 1 (ENSG00000145416), score: 0.56 MARCH4membrane-associated ring finger (C3HC4) 4 (ENSG00000144583), score: 0.56 MARCKSL1MARCKS-like 1 (ENSG00000175130), score: 0.71 MAST3microtubule associated serine/threonine kinase 3 (ENSG00000099308), score: 0.6 MATN1matrilin 1, cartilage matrix protein (ENSG00000162510), score: 0.64 MED10mediator complex subunit 10 (ENSG00000133398), score: 0.55 MED20mediator complex subunit 20 (ENSG00000124641), score: -0.59 MEGF10multiple EGF-like-domains 10 (ENSG00000145794), score: 0.56 MID1IP1MID1 interacting protein 1 (gastrulation specific G12 homolog (zebrafish)) (ENSG00000165175), score: 0.6 MKRN3makorin ring finger protein 3 (ENSG00000179455), score: 0.72 MMP17matrix metallopeptidase 17 (membrane-inserted) (ENSG00000198598), score: 0.58 MOBKL1BMOB1, Mps One Binder kinase activator-like 1B (yeast) (ENSG00000114978), score: -0.55 MOGmyelin oligodendrocyte glycoprotein (ENSG00000204655), score: 0.73 MT3metallothionein 3 (ENSG00000087250), score: 0.54 MYBBP1AMYB binding protein (P160) 1a (ENSG00000132382), score: 0.58 MYNNmyoneurin (ENSG00000085274), score: -0.55 MYO9Bmyosin IXB (ENSG00000099331), score: 0.65 NANOS3nanos homolog 3 (Drosophila) (ENSG00000187556), score: 0.6 NANPN-acetylneuraminic acid phosphatase (ENSG00000170191), score: -0.61 NCANneurocan (ENSG00000130287), score: 0.54 NEO1neogenin 1 (ENSG00000067141), score: 0.54 NEUROD6neurogenic differentiation 6 (ENSG00000164600), score: 0.54 NINJ2ninjurin 2 (ENSG00000171840), score: 0.55 NIPA1non imprinted in Prader-Willi/Angelman syndrome 1 (ENSG00000170113), score: 0.67 NIPAL4NIPA-like domain containing 4 (ENSG00000172548), score: 1 NKX2-2NK2 homeobox 2 (ENSG00000125820), score: 0.74 NLKnemo-like kinase (ENSG00000087095), score: 0.54 NPAS1neuronal PAS domain protein 1 (ENSG00000130751), score: 0.56 NPC1Niemann-Pick disease, type C1 (ENSG00000141458), score: 0.84 NTN5netrin 5 (ENSG00000142233), score: 0.54 NTNG2netrin G2 (ENSG00000196358), score: 0.65 NTSR2neurotensin receptor 2 (ENSG00000169006), score: 0.57 OLIG2oligodendrocyte lineage transcription factor 2 (ENSG00000205927), score: 0.69 OPALINoligodendrocytic myelin paranodal and inner loop protein (ENSG00000197430), score: 0.74 OR2T6olfactory receptor, family 2, subfamily T, member 6 (ENSG00000198104), score: 0.82 P2RX7purinergic receptor P2X, ligand-gated ion channel, 7 (ENSG00000089041), score: 0.7 PAAF1proteasomal ATPase-associated factor 1 (ENSG00000175575), score: 0.59 PACS2phosphofurin acidic cluster sorting protein 2 (ENSG00000179364), score: 0.64 PADI2peptidyl arginine deiminase, type II (ENSG00000117115), score: 0.57 PAQR6progestin and adipoQ receptor family member VI (ENSG00000160781), score: 0.7 PCDH10protocadherin 10 (ENSG00000138650), score: 0.56 PCDH9protocadherin 9 (ENSG00000184226), score: 0.53 PDE6Bphosphodiesterase 6B, cGMP-specific, rod, beta (ENSG00000133256), score: 0.56 PEX13peroxisomal biogenesis factor 13 (ENSG00000162928), score: -0.59 PEX5Lperoxisomal biogenesis factor 5-like (ENSG00000114757), score: 0.54 PGM2phosphoglucomutase 2 (ENSG00000169299), score: -0.63 PHLDA3pleckstrin homology-like domain, family A, member 3 (ENSG00000174307), score: 0.6 PHLDB1pleckstrin homology-like domain, family B, member 1 (ENSG00000019144), score: 0.54 PHLPP1PH domain and leucine rich repeat protein phosphatase 1 (ENSG00000081913), score: 0.64 PIP4K2Aphosphatidylinositol-5-phosphate 4-kinase, type II, alpha (ENSG00000150867), score: 0.77 PIP5KL1phosphatidylinositol-4-phosphate 5-kinase-like 1 (ENSG00000167103), score: 0.57 PKMYT1protein kinase, membrane associated tyrosine/threonine 1 (ENSG00000127564), score: 0.63 PLA2G4Cphospholipase A2, group IVC (cytosolic, calcium-independent) (ENSG00000105499), score: 0.54 PLEKHB1pleckstrin homology domain containing, family B (evectins) member 1 (ENSG00000021300), score: 0.64 PLEKHG3pleckstrin homology domain containing, family G (with RhoGef domain) member 3 (ENSG00000126822), score: 0.65 PLEKHH1pleckstrin homology domain containing, family H (with MyTH4 domain) member 1 (ENSG00000054690), score: 0.64 PLLPplasmolipin (ENSG00000102934), score: 0.66 PLP1proteolipid protein 1 (ENSG00000123560), score: 0.65 PLXNB3plexin B3 (ENSG00000198753), score: 0.7 PMP2peripheral myelin protein 2 (ENSG00000147588), score: 0.67 PMP22peripheral myelin protein 22 (ENSG00000109099), score: 0.63 PNCKpregnancy up-regulated non-ubiquitously expressed CaM kinase (ENSG00000130822), score: 0.59 PNMAL2PNMA-like 2 (ENSG00000204851), score: 0.55 PNPLA6patatin-like phospholipase domain containing 6 (ENSG00000032444), score: 0.59 PPP1R14Aprotein phosphatase 1, regulatory (inhibitor) subunit 14A (ENSG00000167641), score: 0.72 PRAF2PRA1 domain family, member 2 (ENSG00000243279), score: 0.66 PREX1phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 1 (ENSG00000124126), score: 0.72 PRIMA1proline rich membrane anchor 1 (ENSG00000175785), score: 0.57 PRKCQprotein kinase C, theta (ENSG00000065675), score: 0.7 PRMT2protein arginine methyltransferase 2 (ENSG00000160310), score: 0.53 PRPF18PRP18 pre-mRNA processing factor 18 homolog (S. cerevisiae) (ENSG00000165630), score: -0.69 PRR18proline rich 18 (ENSG00000176381), score: 0.86 PRR5Lproline rich 5 like (ENSG00000135362), score: 0.68 PRR7proline rich 7 (synaptic) (ENSG00000131188), score: 0.63 PRRC1proline-rich coiled-coil 1 (ENSG00000164244), score: -0.58 PRRG1proline rich Gla (G-carboxyglutamic acid) 1 (ENSG00000130962), score: 0.57 PSRC1proline/serine-rich coiled-coil 1 (ENSG00000134222), score: 0.68 PTK2PTK2 protein tyrosine kinase 2 (ENSG00000169398), score: 0.78 PTPRHprotein tyrosine phosphatase, receptor type, H (ENSG00000080031), score: 0.75 PXKPX domain containing serine/threonine kinase (ENSG00000168297), score: 0.67 QDPRquinoid dihydropteridine reductase (ENSG00000151552), score: 0.54 RAB11BRAB11B, member RAS oncogene family (ENSG00000185236), score: 0.58 RAB27ARAB27A, member RAS oncogene family (ENSG00000069974), score: -0.55 RAB40BRAB40B, member RAS oncogene family (ENSG00000141542), score: 0.65 RAB4BRAB4B, member RAS oncogene family (ENSG00000167578), score: 0.58 RALGDSral guanine nucleotide dissociation stimulator (ENSG00000160271), score: 0.63 RANGAP1Ran GTPase activating protein 1 (ENSG00000100401), score: 0.56 RAPGEF5Rap guanine nucleotide exchange factor (GEF) 5 (ENSG00000136237), score: 0.66 RARSarginyl-tRNA synthetase (ENSG00000113643), score: -0.57 RASA3RAS p21 protein activator 3 (ENSG00000185989), score: 0.53 RASAL1RAS protein activator like 1 (GAP1 like) (ENSG00000111344), score: 0.67 RASL10ARAS-like, family 10, member A (ENSG00000100276), score: 0.56 RASSF2Ras association (RalGDS/AF-6) domain family member 2 (ENSG00000101265), score: 0.59 RBP3retinol binding protein 3, interstitial (ENSG00000107618), score: 0.58 RHBDD2rhomboid domain containing 2 (ENSG00000005486), score: 0.58 RHBDL2rhomboid, veinlet-like 2 (Drosophila) (ENSG00000158315), score: 0.63 RHOGras homolog gene family, member G (rho G) (ENSG00000177105), score: 0.65 RHOUras homolog gene family, member U (ENSG00000116574), score: 0.64 RNF220ring finger protein 220 (ENSG00000187147), score: 0.67 ROM1retinal outer segment membrane protein 1 (ENSG00000149489), score: 0.67 RPF1ribosome production factor 1 homolog (S. cerevisiae) (ENSG00000117133), score: -0.58 RPRMreprimo, TP53 dependent G2 arrest mediator candidate (ENSG00000177519), score: 0.57 RRM1ribonucleotide reductase M1 (ENSG00000167325), score: -0.59 RTBDNretbindin (ENSG00000132026), score: 0.62 RTN3reticulon 3 (ENSG00000133318), score: 0.53 SAMD4Bsterile alpha motif domain containing 4B (ENSG00000179134), score: 0.63 SBF1SET binding factor 1 (ENSG00000100241), score: 0.55 SCN3Bsodium channel, voltage-gated, type III, beta (ENSG00000166257), score: 0.55 SCYL2SCY1-like 2 (S. cerevisiae) (ENSG00000136021), score: -0.57 SEC14L5SEC14-like 5 (S. cerevisiae) (ENSG00000103184), score: 0.66 SEMA3Bsema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3B (ENSG00000012171), score: 0.56 SEMA4Dsema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4D (ENSG00000187764), score: 0.64 SEMA6Asema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6A (ENSG00000092421), score: 0.68 SEPT4septin 4 (ENSG00000108387), score: 0.56 SEPT8septin 8 (ENSG00000164402), score: 0.55 SERPINI1serpin peptidase inhibitor, clade I (neuroserpin), member 1 (ENSG00000163536), score: 0.54 SETD6SET domain containing 6 (ENSG00000103037), score: -0.56 SH3GLB2SH3-domain GRB2-like endophilin B2 (ENSG00000148341), score: 0.67 SH3TC2SH3 domain and tetratricopeptide repeats 2 (ENSG00000169247), score: 0.69 SHC4SHC (Src homology 2 domain containing) family, member 4 (ENSG00000185634), score: 0.6 SHISA4shisa homolog 4 (Xenopus laevis) (ENSG00000198892), score: 0.56 SIRT2sirtuin 2 (ENSG00000068903), score: 0.85 SLAIN1SLAIN motif family, member 1 (ENSG00000139737), score: 0.67 SLC22A15solute carrier family 22, member 15 (ENSG00000163393), score: 0.56 SLC24A2solute carrier family 24 (sodium/potassium/calcium exchanger), member 2 (ENSG00000155886), score: 0.56 SLC25A29solute carrier family 25, member 29 (ENSG00000197119), score: 0.54 SLC31A2solute carrier family 31 (copper transporters), member 2 (ENSG00000136867), score: 0.7 SLC44A1solute carrier family 44, member 1 (ENSG00000070214), score: 0.67 SLC5A11solute carrier family 5 (sodium/glucose cotransporter), member 11 (ENSG00000158865), score: 0.8 SLC6A9solute carrier family 6 (neurotransmitter transporter, glycine), member 9 (ENSG00000196517), score: 0.67 SLCO1A2solute carrier organic anion transporter family, member 1A2 (ENSG00000084453), score: 0.63 SLIT1slit homolog 1 (Drosophila) (ENSG00000187122), score: 0.57 SLITRK1SLIT and NTRK-like family, member 1 (ENSG00000178235), score: 0.57 SMARCA1SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 1 (ENSG00000102038), score: -0.56 SMARCE1SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily e, member 1 (ENSG00000073584), score: -0.55 SMEK1SMEK homolog 1, suppressor of mek1 (Dictyostelium) (ENSG00000100796), score: -0.6 SMOXspermine oxidase (ENSG00000088826), score: 0.55 SNAI3snail homolog 3 (Drosophila) (ENSG00000185669), score: 0.64 SNX2sorting nexin 2 (ENSG00000205302), score: -0.57 SOHLH1spermatogenesis and oogenesis specific basic helix-loop-helix 1 (ENSG00000165643), score: 0.59 SORCS2sortilin-related VPS10 domain containing receptor 2 (ENSG00000184985), score: 0.53 SOX10SRY (sex determining region Y)-box 10 (ENSG00000100146), score: 0.72 SOX8SRY (sex determining region Y)-box 8 (ENSG00000005513), score: 0.75 SP3Sp3 transcription factor (ENSG00000172845), score: -0.6 SPRED1sprouty-related, EVH1 domain containing 1 (ENSG00000166068), score: 0.56 SPTY2D1SPT2, Suppressor of Ty, domain containing 1 (S. cerevisiae) (ENSG00000179119), score: -0.59 SRFBP1serum response factor binding protein 1 (ENSG00000151304), score: -0.55 SSTsomatostatin (ENSG00000157005), score: 0.62 STK32Cserine/threonine kinase 32C (ENSG00000165752), score: 0.58 STOML1stomatin (EPB72)-like 1 (ENSG00000067221), score: 0.61 SYNJ2synaptojanin 2 (ENSG00000078269), score: 0.6 SYT15synaptotagmin XV (ENSG00000204176), score: 0.64 SYT5synaptotagmin V (ENSG00000129990), score: 0.54 TBC1D12TBC1 domain family, member 12 (ENSG00000108239), score: 0.54 TBCBtubulin folding cofactor B (ENSG00000105254), score: 0.66 TGS1trimethylguanosine synthase 1 (ENSG00000137574), score: -0.57 THEMISthymocyte selection associated (ENSG00000172673), score: 0.6 THRAP3thyroid hormone receptor associated protein 3 (ENSG00000054118), score: -0.58 TJAP1tight junction associated protein 1 (peripheral) (ENSG00000137221), score: 0.77 TMC6transmembrane channel-like 6 (ENSG00000141524), score: 0.65 TMCC2transmembrane and coiled-coil domain family 2 (ENSG00000133069), score: 0.54 TMCC3transmembrane and coiled-coil domain family 3 (ENSG00000057704), score: 0.58 TMEFF2transmembrane protein with EGF-like and two follistatin-like domains 2 (ENSG00000144339), score: 0.58 TMEM144transmembrane protein 144 (ENSG00000164124), score: 0.82 TMEM151Atransmembrane protein 151A (ENSG00000179292), score: 0.68 TMEM160transmembrane protein 160 (ENSG00000130748), score: 0.59 TMEM184Btransmembrane protein 184B (ENSG00000198792), score: 0.59 TMEM54transmembrane protein 54 (ENSG00000121900), score: 0.56 TMEM55Btransmembrane protein 55B (ENSG00000165782), score: 0.53 TMEM66transmembrane protein 66 (ENSG00000133872), score: 0.54 TMIEtransmembrane inner ear (ENSG00000181585), score: 0.58 TMTC2transmembrane and tetratricopeptide repeat containing 2 (ENSG00000179104), score: 0.57 TNFRSF13Ctumor necrosis factor receptor superfamily, member 13C (ENSG00000159958), score: 0.57 TP53INP2tumor protein p53 inducible nuclear protein 2 (ENSG00000078804), score: 0.76 TPPPtubulin polymerization promoting protein (ENSG00000171368), score: 0.56 TREM2triggering receptor expressed on myeloid cells 2 (ENSG00000095970), score: 0.64 TRIM41tripartite motif-containing 41 (ENSG00000146063), score: 0.55 TRIP11thyroid hormone receptor interactor 11 (ENSG00000100815), score: -0.55 TRMT61AtRNA methyltransferase 61 homolog A (S. cerevisiae) (ENSG00000166166), score: 0.58 TRPM2transient receptor potential cation channel, subfamily M, member 2 (ENSG00000142185), score: 0.59 TRPM7transient receptor potential cation channel, subfamily M, member 7 (ENSG00000092439), score: -0.56 TSPAN15tetraspanin 15 (ENSG00000099282), score: 0.62 UGT8UDP glycosyltransferase 8 (ENSG00000174607), score: 0.72 UNC5Cunc-5 homolog C (C. elegans) (ENSG00000182168), score: 0.62 USP3ubiquitin specific peptidase 3 (ENSG00000140455), score: -0.58 USP53ubiquitin specific peptidase 53 (ENSG00000145390), score: -0.6 USP54ubiquitin specific peptidase 54 (ENSG00000166348), score: 0.56 VAMP1vesicle-associated membrane protein 1 (synaptobrevin 1) (ENSG00000139190), score: 0.55 VGFVGF nerve growth factor inducible (ENSG00000128564), score: 0.55 VSTM2BV-set and transmembrane domain containing 2B (ENSG00000187135), score: 0.64 WDR3WD repeat domain 3 (ENSG00000065183), score: -0.55 WDR33WD repeat domain 33 (ENSG00000136709), score: -0.59 WDR43WD repeat domain 43 (ENSG00000163811), score: -0.55 WEE1WEE1 homolog (S. pombe) (ENSG00000166483), score: -0.57 WNT10Bwingless-type MMTV integration site family, member 10B (ENSG00000169884), score: 0.57 WRBtryptophan rich basic protein (ENSG00000182093), score: 0.53 YME1L1YME1-like 1 (S. cerevisiae) (ENSG00000136758), score: -0.59 ZCCHC24zinc finger, CCHC domain containing 24 (ENSG00000165424), score: 0.61 ZEB2zinc finger E-box binding homeobox 2 (ENSG00000169554), score: 0.6 ZFP57zinc finger protein 57 homolog (mouse) (ENSG00000204644), score: 0.71 ZNF536zinc finger protein 536 (ENSG00000198597), score: 0.61
| Id | species | tissue | sex | individual |
|---|---|---|---|---|
| ptr_br_m5_ca1 | ptr | br | m | 5 |
| ptr_br_m2_ca1 | ptr | br | m | 2 |
| ptr_br_m3_ca1 | ptr | br | m | 3 |
| hsa_br_f_ca1 | hsa | br | f | _ |
| hsa_br_m7_ca1 | hsa | br | m | 7 |
| hsa_br_m3_ca1 | hsa | br | m | 3 |
| hsa_br_m6_ca1 | hsa | br | m | 6 |