Difference between revisions of "Identification of obesity and BMI associated intergenic long noncoding RNAs"
Sbprm2015 4 (talk | contribs) |
|||
| (4 intermediate revisions by 3 users not shown) | |||
| Line 1: | Line 1: | ||
| − | ''' | + | '''Introduction''' |
| + | Long intergenic non-coding RNA is an ARN type longer than 200 nucleotides. They are not translated but a minor part of them seems to interact with some cellular processes. Among these processes we can point out the regulation and the modulation of gene expressions. The lincRNA are often tissue specific. | ||
| − | ''' | + | '''Goals of the project:''' |
| + | Establishing if a relation between some specific LincRNAs expression levels and particular genetic polymorphisms; recently linked to diseases traits through Genome Wide Association Studies, GWAS. | ||
| − | + | This project led our work on SNPs linked to auto-immune traits and particularly auto-immune diseases. | |
| − | '''Supervisors:'''[[User: | + | '''Dataset and methodolgy:''' |
| + | During the whole assignment, the data used for the lincRNAs expression levels and the SNPs came from a study; published in September 2013 in Nature . These data are based on the sequencing of lymphoid cells lines of 462 individuals wherein we selected only the European individuals in order to maintain consistency with the data of SNPs related traits. Only the SNPs associated to an auto-immune trait with a p-value inferior to 5×10-8 were selected to the usual standard of GWAS studies. | ||
| + | |||
| + | The other part of the data used in this project comes from the GWAS catalogue and were constituted of 579 SNPs linked to some auto-immune diseases i.e. | ||
| + | |||
| + | - Hypothyroidism | ||
| + | - Multiple sclerosis | ||
| + | - Psoriatic arthritis | ||
| + | - Rheumatoid arthritis | ||
| + | - Systemic lupus erythematosus and systemic sclerosis | ||
| + | - Type 1 diabetes | ||
| + | |||
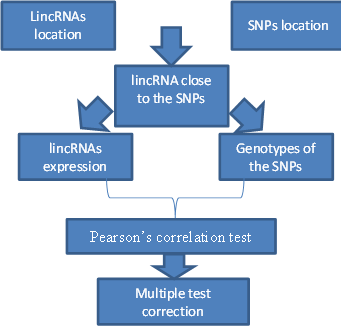
| + | As the project involves a cis-eQTL analysis, the first step in the data’s manipulation was to select the LincRNAs located close to the SNPs (+/-10-6 bp). Then to initiate the research of a correlation; a multiple test with a permutation correction per was employed. The permutation correction allows us to maintain the false positive-result or False Discover Rate fewer than 10%. | ||
| + | |||
| + | The methodology and the employment of the data can be summarized by this schema: | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | [[File:methodolgy.png]] | ||
| + | |||
| + | |||
| + | '''Results:''' | ||
| + | After the application of the method exposed upper the following results were obtained: | ||
| + | |||
| + | |||
| + | [[File:resultats.png]] | ||
| + | |||
| + | [[File:tab.png]] | ||
| + | |||
| + | |||
| + | |||
| + | |||
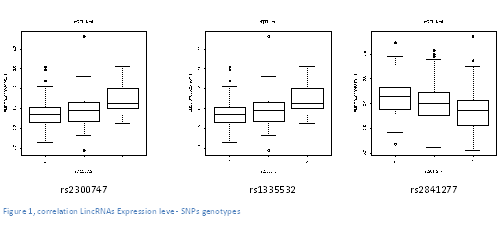
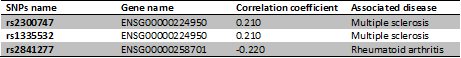
| + | The first and the second correlation exposed, concerns the same lincRNA (gene ENSG00000224950) but two different SNPs (rs2300747 & rs1335532) linked to the multiple sclerosis, a disease which damages the myelin around the cellular cells, producing mental and physical problems. These two correlations have therefore the same value, they are both positives too. | ||
| + | |||
| + | The result of the third correlation was negative and stays between the LincRNA (gene ENSG00000258701) and the SNPs rs2841277. This third SNPs is associated the Rheumatoid arthritis, a chronic inflammatory disorder that affects joints in the articulations. | ||
| + | |||
| + | '''Discussion and prospective:''' | ||
| + | To obtain significant results; a 10% FDR had been used instead of the usual 5%. It increased the risk of false positives results and reduced the test precision but allowed the highlight of a probable correlation between the expression of some lincRNAs and auto-immune traits. | ||
| + | |||
| + | Better results could have obtained by testing other tissues than LCL. For example it could be interesting to apply the methodology of this project to tissues like skin, neurone or bones which are also related to the diseases exposed in this assignment | ||
| + | |||
| + | |||
| + | '''Conclusion:''' | ||
| + | The lincRNAs are today still not entirely understood. There is probably a lot more to discover about the lincRNAs and their functions or interactions with some biological processes. That is why lincRNA will probably the subject of many research in the next years. | ||
| + | |||
| + | '''Students:''' | ||
| + | Eric Gähwiler | ||
| + | Karim Hamidi | ||
| + | Virginie Ricci | ||
| + | |||
| + | '''Supervisors:''' [[User: JenniferTan|Jennifer Tan]] and [[User: AnaMarques|Ana Claudia Machado Rebelo Marques]] | ||
| + | |||
| + | [[Media:EQTL-Obesity-JenniferTan.pptx]] | ||
| + | [[Media:Présentation finale.pptx]] | ||
Latest revision as of 11:09, 29 May 2015
Introduction Long intergenic non-coding RNA is an ARN type longer than 200 nucleotides. They are not translated but a minor part of them seems to interact with some cellular processes. Among these processes we can point out the regulation and the modulation of gene expressions. The lincRNA are often tissue specific.
Goals of the project: Establishing if a relation between some specific LincRNAs expression levels and particular genetic polymorphisms; recently linked to diseases traits through Genome Wide Association Studies, GWAS.
This project led our work on SNPs linked to auto-immune traits and particularly auto-immune diseases.
Dataset and methodolgy: During the whole assignment, the data used for the lincRNAs expression levels and the SNPs came from a study; published in September 2013 in Nature . These data are based on the sequencing of lymphoid cells lines of 462 individuals wherein we selected only the European individuals in order to maintain consistency with the data of SNPs related traits. Only the SNPs associated to an auto-immune trait with a p-value inferior to 5×10-8 were selected to the usual standard of GWAS studies.
The other part of the data used in this project comes from the GWAS catalogue and were constituted of 579 SNPs linked to some auto-immune diseases i.e.
- Hypothyroidism - Multiple sclerosis - Psoriatic arthritis - Rheumatoid arthritis - Systemic lupus erythematosus and systemic sclerosis - Type 1 diabetes
As the project involves a cis-eQTL analysis, the first step in the data’s manipulation was to select the LincRNAs located close to the SNPs (+/-10-6 bp). Then to initiate the research of a correlation; a multiple test with a permutation correction per was employed. The permutation correction allows us to maintain the false positive-result or False Discover Rate fewer than 10%.
The methodology and the employment of the data can be summarized by this schema:
Results:
After the application of the method exposed upper the following results were obtained:
The first and the second correlation exposed, concerns the same lincRNA (gene ENSG00000224950) but two different SNPs (rs2300747 & rs1335532) linked to the multiple sclerosis, a disease which damages the myelin around the cellular cells, producing mental and physical problems. These two correlations have therefore the same value, they are both positives too.
The result of the third correlation was negative and stays between the LincRNA (gene ENSG00000258701) and the SNPs rs2841277. This third SNPs is associated the Rheumatoid arthritis, a chronic inflammatory disorder that affects joints in the articulations.
Discussion and prospective: To obtain significant results; a 10% FDR had been used instead of the usual 5%. It increased the risk of false positives results and reduced the test precision but allowed the highlight of a probable correlation between the expression of some lincRNAs and auto-immune traits.
Better results could have obtained by testing other tissues than LCL. For example it could be interesting to apply the methodology of this project to tissues like skin, neurone or bones which are also related to the diseases exposed in this assignment
Conclusion:
The lincRNAs are today still not entirely understood. There is probably a lot more to discover about the lincRNAs and their functions or interactions with some biological processes. That is why lincRNA will probably the subject of many research in the next years.
Students: Eric Gähwiler Karim Hamidi Virginie Ricci
Supervisors: Jennifer Tan and Ana Claudia Machado Rebelo Marques
Media:EQTL-Obesity-JenniferTan.pptx Media:Présentation finale.pptx