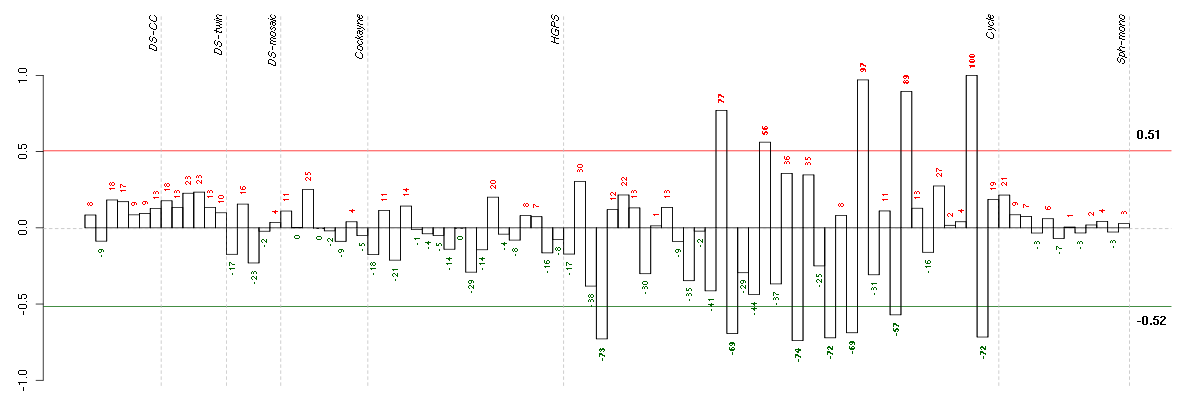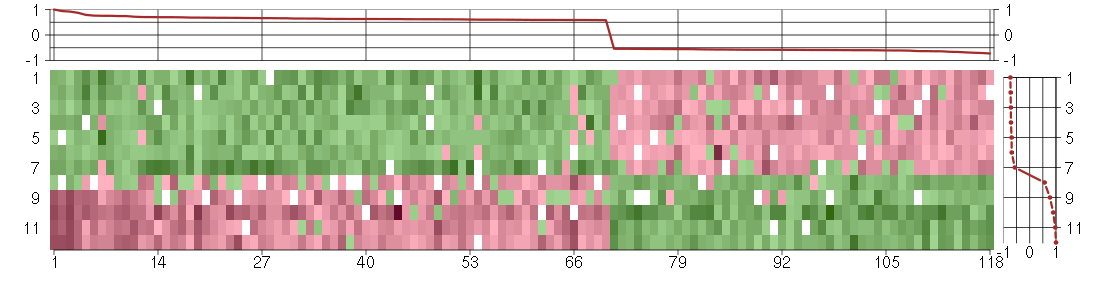

Under-expression is coded with green,
over-expression with red color.


intracellular
The living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
ribonucleoprotein complex
A macromolecular complex containing both protein and RNA molecules.
cytosol
That part of the cytoplasm that does not contain membranous or particulate subcellular components.
ribosome
An intracellular organelle, about 200 A in diameter, consisting of RNA and protein. It is the site of protein biosynthesis resulting from translation of messenger RNA (mRNA). It consists of two subunits, one large and one small, each containing only protein and RNA. Both the ribosome and its subunits are characterized by their sedimentation coefficients, expressed in Svedberg units (symbol: S). Hence, the prokaryotic ribosome (70S) comprises a large (50S) subunit and a small (30S) subunit, while the eukaryotic ribosome (80S) comprises a large (60S) subunit and a small (40S) subunit. Two sites on the ribosomal large subunit are involved in translation, namely the aminoacyl site (A site) and peptidyl site (P site). Ribosomes from prokaryotes, eukaryotes, mitochondria, and chloroplasts have characteristically distinct ribosomal proteins.
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
cell
The basic structural and functional unit of all organisms. Includes the plasma membrane and any external encapsulating structures such as the cell wall and cell envelope.
cytoplasm
All of the contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures.
cytosolic ribosome
A ribosome that is found in the cytosol of the cell. The cytosol is that part of the cytoplasm that does not contain membranous or particulate subcellular components.
macromolecular complex
A stable assembly of two or more macromolecules, i.e. proteins, nucleic acids, carbohydrates or lipids, in which the constituent parts function together.
organelle
Organized structure of distinctive morphology and function. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton. Excludes the plasma membrane.
non-membrane-bounded organelle
Organized structure of distinctive morphology and function, not bounded by a lipid bilayer membrane. Includes ribosomes, the cytoskeleton and chromosomes.
intracellular organelle
Organized structure of distinctive morphology and function, occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton. Excludes the plasma membrane.
intracellular non-membrane-bounded organelle
Organized structure of distinctive morphology and function, not bounded by a lipid bilayer membrane and occurring within the cell. Includes ribosomes, the cytoskeleton and chromosomes.
intracellular part
Any constituent part of the living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
cytoplasmic part
Any constituent part of the cytoplasm, all of the contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures.
cytosolic part
Any constituent part of cytosol, that part of the cytoplasm that does not contain membranous or particulate subcellular components.
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
all
This term is the most general term possible
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
intracellular non-membrane-bounded organelle
Organized structure of distinctive morphology and function, not bounded by a lipid bilayer membrane and occurring within the cell. Includes ribosomes, the cytoskeleton and chromosomes.
intracellular part
Any constituent part of the living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
ribonucleoprotein complex
A macromolecular complex containing both protein and RNA molecules.
intracellular organelle
Organized structure of distinctive morphology and function, occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton. Excludes the plasma membrane.
ribosome
An intracellular organelle, about 200 A in diameter, consisting of RNA and protein. It is the site of protein biosynthesis resulting from translation of messenger RNA (mRNA). It consists of two subunits, one large and one small, each containing only protein and RNA. Both the ribosome and its subunits are characterized by their sedimentation coefficients, expressed in Svedberg units (symbol: S). Hence, the prokaryotic ribosome (70S) comprises a large (50S) subunit and a small (30S) subunit, while the eukaryotic ribosome (80S) comprises a large (60S) subunit and a small (40S) subunit. Two sites on the ribosomal large subunit are involved in translation, namely the aminoacyl site (A site) and peptidyl site (P site). Ribosomes from prokaryotes, eukaryotes, mitochondria, and chloroplasts have characteristically distinct ribosomal proteins.
cytoplasmic part
Any constituent part of the cytoplasm, all of the contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures.
ribosome
An intracellular organelle, about 200 A in diameter, consisting of RNA and protein. It is the site of protein biosynthesis resulting from translation of messenger RNA (mRNA). It consists of two subunits, one large and one small, each containing only protein and RNA. Both the ribosome and its subunits are characterized by their sedimentation coefficients, expressed in Svedberg units (symbol: S). Hence, the prokaryotic ribosome (70S) comprises a large (50S) subunit and a small (30S) subunit, while the eukaryotic ribosome (80S) comprises a large (60S) subunit and a small (40S) subunit. Two sites on the ribosomal large subunit are involved in translation, namely the aminoacyl site (A site) and peptidyl site (P site). Ribosomes from prokaryotes, eukaryotes, mitochondria, and chloroplasts have characteristically distinct ribosomal proteins.
cytosolic part
Any constituent part of cytosol, that part of the cytoplasm that does not contain membranous or particulate subcellular components.
cytosolic ribosome
A ribosome that is found in the cytosol of the cell. The cytosol is that part of the cytoplasm that does not contain membranous or particulate subcellular components.

ACANaggrecan (205679_x_at), score: 0.63 ADCK2aarF domain containing kinase 2 (221893_s_at), score: -0.7 ADRB2adrenergic, beta-2-, receptor, surface (206170_at), score: -0.61 ALDH3B1aldehyde dehydrogenase 3 family, member B1 (211004_s_at), score: 0.63 ALDH4A1aldehyde dehydrogenase 4 family, member A1 (211552_s_at), score: 0.59 ANKRD28ankyrin repeat domain 28 (213035_at), score: 0.6 ANXA10annexin A10 (210143_at), score: -0.64 APOL2apolipoprotein L, 2 (221653_x_at), score: 0.58 ASB1ankyrin repeat and SOCS box-containing 1 (212819_at), score: -0.61 B4GALT1UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 1 (201883_s_at), score: 0.74 BCANbrevican (91920_at), score: 0.63 BTN3A1butyrophilin, subfamily 3, member A1 (209770_at), score: 0.66 C1orf66chromosome 1 open reading frame 66 (218914_at), score: 0.62 C6orf155chromosome 6 open reading frame 155 (220324_at), score: -0.58 C6orf62chromosome 6 open reading frame 62 (213875_x_at), score: 0.94 C7orf58chromosome 7 open reading frame 58 (220032_at), score: -0.61 C9orf114chromosome 9 open reading frame 114 (218565_at), score: 0.62 CAPZA1capping protein (actin filament) muscle Z-line, alpha 1 (208374_s_at), score: 0.63 CCDC28Acoiled-coil domain containing 28A (209479_at), score: 0.59 CCDC94coiled-coil domain containing 94 (204335_at), score: 0.66 CCNE1cyclin E1 (213523_at), score: -0.59 CDCA4cell division cycle associated 4 (218399_s_at), score: -0.58 CDCP1CUB domain containing protein 1 (218451_at), score: -0.55 CENPTcentromere protein T (218148_at), score: -0.6 CHPcalcium binding protein P22 (214665_s_at), score: 0.69 CNNM2cyclin M2 (206818_s_at), score: 0.61 CORO2Bcoronin, actin binding protein, 2B (209789_at), score: -0.71 CPA4carboxypeptidase A4 (205832_at), score: 0.6 CTTNcortactin (201059_at), score: 0.76 DCAKDdephospho-CoA kinase domain containing (221224_s_at), score: 0.66 DDIT3DNA-damage-inducible transcript 3 (209383_at), score: 0.63 DIRAS3DIRAS family, GTP-binding RAS-like 3 (215506_s_at), score: -0.54 DOK4docking protein 4 (209691_s_at), score: -0.64 DSN1DSN1, MIND kinetochore complex component, homolog (S. cerevisiae) (219512_at), score: -0.6 DSPPdentin sialophosphoprotein (221681_s_at), score: 0.64 DUSP5dual specificity phosphatase 5 (209457_at), score: -0.58 EIF4Beukaryotic translation initiation factor 4B (219599_at), score: 0.66 EIF4EBP2eukaryotic translation initiation factor 4E binding protein 2 (208770_s_at), score: -0.6 EML1echinoderm microtubule associated protein like 1 (204797_s_at), score: -0.54 FAM117Afamily with sequence similarity 117, member A (221249_s_at), score: 0.58 FAM70Afamily with sequence similarity 70, member A (219895_at), score: -0.57 FAM86Cfamily with sequence similarity 86, member C (220353_at), score: 0.68 FEM1Cfem-1 homolog c (C. elegans) (213341_at), score: -0.59 FZD4frizzled homolog 4 (Drosophila) (218665_at), score: 0.59 GALNSgalactosamine (N-acetyl)-6-sulfate sulfatase (206335_at), score: 0.6 GCHFRGTP cyclohydrolase I feedback regulator (204867_at), score: 0.63 GPR144G protein-coupled receptor 144 (216289_at), score: 0.75 GRB14growth factor receptor-bound protein 14 (206204_at), score: -0.54 HAB1B1 for mucin (215778_x_at), score: 0.6 HINFPhistone H4 transcription factor (206495_s_at), score: 0.64 HLA-DQB1major histocompatibility complex, class II, DQ beta 1 (211654_x_at), score: 0.59 IL13RA2interleukin 13 receptor, alpha 2 (206172_at), score: -0.56 IL33interleukin 33 (209821_at), score: -0.63 ITGALintegrin, alpha L (antigen CD11A (p180), lymphocyte function-associated antigen 1; alpha polypeptide) (213475_s_at), score: 0.79 KRT33Akeratin 33A (208483_x_at), score: -0.58 LILRB3leukocyte immunoglobulin-like receptor, subfamily B (with TM and ITIM domains), member 3 (211133_x_at), score: 0.68 LOC149478hypothetical protein LOC149478 (215462_at), score: 0.63 LOC388152hypothetical LOC388152 (220602_s_at), score: 0.7 MAN1B1mannosidase, alpha, class 1B, member 1 (65884_at), score: -0.59 MAP1Smicrotubule-associated protein 1S (218522_s_at), score: -0.63 MARCH3membrane-associated ring finger (C3HC4) 3 (213256_at), score: -0.56 MNTMAX binding protein (204206_at), score: 0.59 MOSPD1motile sperm domain containing 1 (218853_s_at), score: 0.61 MUL1mitochondrial E3 ubiquitin ligase 1 (218246_at), score: -0.57 NEDD4Lneural precursor cell expressed, developmentally down-regulated 4-like (212445_s_at), score: -0.59 NFATC2IPnuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 2 interacting protein (217526_at), score: -0.73 NFIXnuclear factor I/X (CCAAT-binding transcription factor) (209807_s_at), score: 0.7 NTHL1nth endonuclease III-like 1 (E. coli) (209731_at), score: 0.7 PCCApropionyl Coenzyme A carboxylase, alpha polypeptide (203860_at), score: 0.61 PDK3pyruvate dehydrogenase kinase, isozyme 3 (221957_at), score: -0.55 PGPEP1pyroglutamyl-peptidase I (219891_at), score: 0.67 PKIAprotein kinase (cAMP-dependent, catalytic) inhibitor alpha (204612_at), score: -0.61 POM121POM121 membrane glycoprotein (rat) (212178_s_at), score: -0.55 PRKG2protein kinase, cGMP-dependent, type II (207505_at), score: -0.6 PRSS3protease, serine, 3 (213421_x_at), score: -0.66 PTPN6protein tyrosine phosphatase, non-receptor type 6 (206687_s_at), score: 0.6 PXNpaxillin (211823_s_at), score: -0.67 RAD54L2RAD54-like 2 (S. cerevisiae) (213205_s_at), score: -0.55 RHOBTB1Rho-related BTB domain containing 1 (212651_at), score: 0.65 RIPK1receptor (TNFRSF)-interacting serine-threonine kinase 1 (209941_at), score: -0.58 RPL14ribosomal protein L14 (219138_at), score: 0.72 RPL18AP6ribosomal protein L18a pseudogene 6 (216383_at), score: 0.67 RPL27Aribosomal protein L27a (212044_s_at), score: 1 RPL38ribosomal protein L38 (202028_s_at), score: 0.87 RPLP2ribosomal protein, large, P2 (200908_s_at), score: 0.74 RPS11ribosomal protein S11 (213350_at), score: 0.92 RPS19ribosomal protein S19 (202648_at), score: 0.61 SAT1spermidine/spermine N1-acetyltransferase 1 (213988_s_at), score: -0.68 SERPINB2serpin peptidase inhibitor, clade B (ovalbumin), member 2 (204614_at), score: -0.54 SETMARSET domain and mariner transposase fusion gene (206554_x_at), score: 0.7 SLC25A16solute carrier family 25 (mitochondrial carrier; Graves disease autoantigen), member 16 (210686_x_at), score: 0.63 SLC29A1solute carrier family 29 (nucleoside transporters), member 1 (201801_s_at), score: -0.59 SLC35F5solute carrier family 35, member F5 (220123_at), score: -0.57 SNHG3small nucleolar RNA host gene 3 (non-protein coding) (215011_at), score: 0.59 SNORA21small nucleolar RNA, H/ACA box 21 (215224_at), score: 0.63 SPRY4sprouty homolog 4 (Drosophila) (221489_s_at), score: -0.55 SSR4signal sequence receptor, delta (translocon-associated protein delta) (201004_at), score: 0.59 SUPV3L1suppressor of var1, 3-like 1 (S. cerevisiae) (212894_at), score: 0.59 SYT1synaptotagmin I (203999_at), score: -0.57 SYT11synaptotagmin XI (209198_s_at), score: 0.68 TAF15TAF15 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 68kDa (202840_at), score: 0.63 TAL1T-cell acute lymphocytic leukemia 1 (206283_s_at), score: 0.65 TMSB4Ythymosin beta 4, Y-linked (206769_at), score: 0.62 TNK2tyrosine kinase, non-receptor, 2 (203839_s_at), score: -0.55 TXNthioredoxin (216609_at), score: 0.75 TYMPthymidine phosphorylase (217497_at), score: 0.7 UCP2uncoupling protein 2 (mitochondrial, proton carrier) (208998_at), score: 0.59 USP47ubiquitin specific peptidase 47 (221518_s_at), score: 0.6 VAMP1vesicle-associated membrane protein 1 (synaptobrevin 1) (213326_at), score: 0.62 VCPIP1valosin containing protein (p97)/p47 complex interacting protein 1 (219810_at), score: 0.68 VEGFBvascular endothelial growth factor B (203683_s_at), score: -0.58 WFDC8WAP four-disulfide core domain 8 (215276_at), score: 0.67 ZDHHC18zinc finger, DHHC-type containing 18 (212860_at), score: -0.58 ZFYVE26zinc finger, FYVE domain containing 26 (37943_at), score: 0.67 ZNF281zinc finger protein 281 (218401_s_at), score: 0.62 ZNF318zinc finger protein 318 (203521_s_at), score: -0.58 ZNF814zinc finger protein 814 (60794_f_at), score: -0.59
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-TABM-263-raw-cel-1515486071.cel | 22 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485711.cel | 4 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486131.cel | 25 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486411.cel | 39 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485951.cel | 16 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486171.cel | 27 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486251.cel | 31 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486011.cel | 19 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485931.cel | 15 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486271.cel | 32 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486191.cel | 28 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486391.cel | 38 | 6 | Cycle | hgu133a2 | none | Cycle 1 |