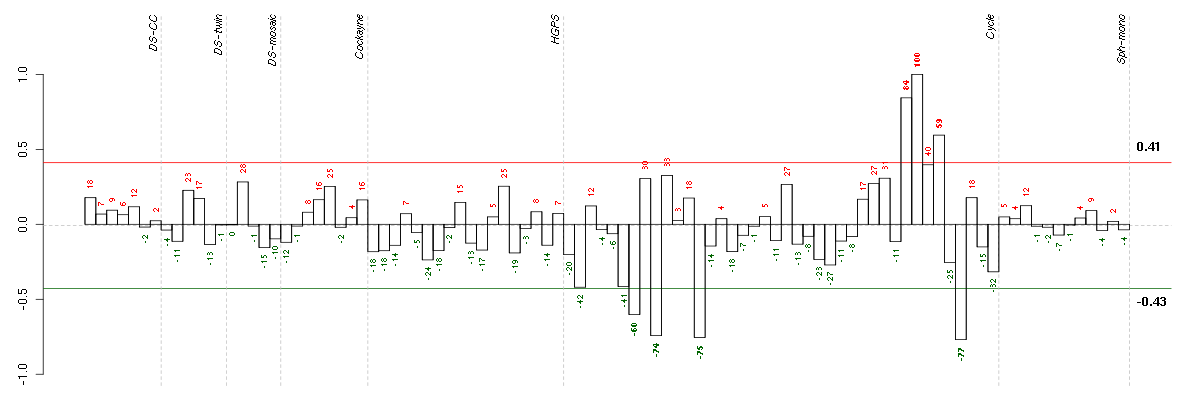

Under-expression is coded with green,
over-expression with red color.

metabolic process
The chemical reactions and pathways, including anabolism and catabolism, by which living organisms transform chemical substances. Metabolic processes typically transform small molecules, but also include macromolecular processes such as DNA repair and replication, and protein synthesis and degradation.
defense response
Reactions, triggered in response to the presence of a foreign body or the occurrence of an injury, which result in restriction of damage to the organism attacked or prevention/recovery from the infection caused by the attack.
nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
The chemical reactions and pathways involving nucleobases, nucleosides, nucleotides and nucleic acids.
transcription
The synthesis of either RNA on a template of DNA or DNA on a template of RNA.
transcription, DNA-dependent
The synthesis of RNA on a template of DNA.
transcription from RNA polymerase II promoter
The synthesis of RNA from a DNA template by RNA polymerase II (Pol II), originating at a Pol II-specific promoter. Includes transcription of messenger RNA (mRNA) and certain small nuclear RNAs (snRNAs).
lipid metabolic process
The chemical reactions and pathways involving lipids, compounds soluble in an organic solvent but not, or sparingly, in an aqueous solvent. Includes fatty acids; neutral fats, other fatty-acid esters, and soaps; long-chain (fatty) alcohols and waxes; sphingoids and other long-chain bases; glycolipids, phospholipids and sphingolipids; and carotenes, polyprenols, sterols, terpenes and other isoprenoids.
isoprenoid metabolic process
The chemical reactions and pathways involving isoprenoid compounds, isoprene (2-methylbuta-1,3-diene) or compounds containing or derived from linked isoprene (3-methyl-2-butenylene) residues.
response to stress
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a disturbance in organismal or cellular homeostasis, usually, but not necessarily, exogenous (e.g. temperature, humidity, ionizing radiation).
cell communication
Any process that mediates interactions between a cell and its surroundings. Encompasses interactions such as signaling or attachment between one cell and another cell, between a cell and an extracellular matrix, or between a cell and any other aspect of its environment.
biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
isoprenoid biosynthetic process
The chemical reactions and pathways resulting in the formation of any isoprenoid compound, isoprene (2-methylbuta-1,3-diene) or compounds containing or derived from linked isoprene (3-methyl-2-butenylene) residues.
lipid biosynthetic process
The chemical reactions and pathways resulting in the formation of lipids, compounds soluble in an organic solvent but not, or sparingly, in an aqueous solvent.
biosynthetic process
The chemical reactions and pathways resulting in the formation of substances; typically the energy-requiring part of metabolism in which simpler substances are transformed into more complex ones.
macromolecule biosynthetic process
The chemical reactions and pathways resulting in the formation of a macromolecule, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass.
cellular process
Any process that is carried out at the cellular level, but not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
gene expression
The process by which a gene's coding sequence is converted into a mature gene product or products (proteins or RNA). This includes the production of an RNA transcript as well as any processing to produce a mature RNA product or an mRNA (for protein-coding genes) and the translation of that mRNA into protein. Some protein processing events may be included when they are required to form an active form of a product from an inactive precursor form.
RNA metabolic process
The chemical reactions and pathways involving RNA, ribonucleic acid, one of the two main type of nucleic acid, consisting of a long, unbranched macromolecule formed from ribonucleotides joined in 3',5'-phosphodiester linkage.
multicellular organismal process
Any biological process, occurring at the level of a multicellular organism, pertinent to its function.
RNA biosynthetic process
The chemical reactions and pathways resulting in the formation of RNA, ribonucleic acid, one of the two main type of nucleic acid, consisting of a long, unbranched macromolecule formed from ribonucleotides joined in 3',5'-phosphodiester linkage. Includes polymerization of ribonucleotide monomers.
cellular macromolecule biosynthetic process
The chemical reactions and pathways resulting in the formation of a macromolecule, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass, carried out by individual cells.
cellular biopolymer metabolic process
The chemical reactions and pathways involving biopolymers, long, repeating chains of monomers found in nature, such as polysaccharides and proteins, as carried out by individual cells.
cellular biopolymer biosynthetic process
The chemical reactions and pathways resulting in the formation of biopolymers, long, repeating chains of monomers found in nature, such as polysaccharides and proteins, as carried out by individual cells.
macromolecule metabolic process
The chemical reactions and pathways involving macromolecules, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass.
biopolymer metabolic process
The chemical reactions and pathways involving biopolymers, long, repeating chains of monomers found in nature, such as polysaccharides and proteins.
biopolymer biosynthetic process
The chemical reactions and pathways resulting in the formation of biopolymers, long, repeating chains of monomers found in nature e.g. polysaccharides and proteins.
cellular metabolic process
The chemical reactions and pathways by which individual cells transform chemical substances.
primary metabolic process
The chemical reactions and pathways involving those compounds which are formed as a part of the normal anabolic and catabolic processes. These processes take place in most, if not all, cells of the organism.
cellular biosynthetic process
The chemical reactions and pathways resulting in the formation of substances, carried out by individual cells.
cellular lipid metabolic process
The chemical reactions and pathways involving lipids, as carried out by individual cells.
cellular macromolecule metabolic process
The chemical reactions and pathways involving macromolecules, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass, as carried out by individual cells.
response to stimulus
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus.
all
This term is the most general term possible
cellular metabolic process
The chemical reactions and pathways by which individual cells transform chemical substances.
macromolecule biosynthetic process
The chemical reactions and pathways resulting in the formation of a macromolecule, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass.
cellular biosynthetic process
The chemical reactions and pathways resulting in the formation of substances, carried out by individual cells.
cellular macromolecule metabolic process
The chemical reactions and pathways involving macromolecules, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass, as carried out by individual cells.
nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
The chemical reactions and pathways involving nucleobases, nucleosides, nucleotides and nucleic acids.
cellular macromolecule biosynthetic process
The chemical reactions and pathways resulting in the formation of a macromolecule, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass, carried out by individual cells.
biopolymer biosynthetic process
The chemical reactions and pathways resulting in the formation of biopolymers, long, repeating chains of monomers found in nature e.g. polysaccharides and proteins.
cellular macromolecule biosynthetic process
The chemical reactions and pathways resulting in the formation of a macromolecule, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass, carried out by individual cells.
cellular biopolymer metabolic process
The chemical reactions and pathways involving biopolymers, long, repeating chains of monomers found in nature, such as polysaccharides and proteins, as carried out by individual cells.
transcription
The synthesis of either RNA on a template of DNA or DNA on a template of RNA.
lipid biosynthetic process
The chemical reactions and pathways resulting in the formation of lipids, compounds soluble in an organic solvent but not, or sparingly, in an aqueous solvent.
cellular lipid metabolic process
The chemical reactions and pathways involving lipids, as carried out by individual cells.
cellular biopolymer biosynthetic process
The chemical reactions and pathways resulting in the formation of biopolymers, long, repeating chains of monomers found in nature, such as polysaccharides and proteins, as carried out by individual cells.
transcription, DNA-dependent
The synthesis of RNA on a template of DNA.
RNA metabolic process
The chemical reactions and pathways involving RNA, ribonucleic acid, one of the two main type of nucleic acid, consisting of a long, unbranched macromolecule formed from ribonucleotides joined in 3',5'-phosphodiester linkage.
cellular biopolymer biosynthetic process
The chemical reactions and pathways resulting in the formation of biopolymers, long, repeating chains of monomers found in nature, such as polysaccharides and proteins, as carried out by individual cells.
RNA biosynthetic process
The chemical reactions and pathways resulting in the formation of RNA, ribonucleic acid, one of the two main type of nucleic acid, consisting of a long, unbranched macromolecule formed from ribonucleotides joined in 3',5'-phosphodiester linkage. Includes polymerization of ribonucleotide monomers.
isoprenoid biosynthetic process
The chemical reactions and pathways resulting in the formation of any isoprenoid compound, isoprene (2-methylbuta-1,3-diene) or compounds containing or derived from linked isoprene (3-methyl-2-butenylene) residues.
transcription
The synthesis of either RNA on a template of DNA or DNA on a template of RNA.


molecular_function
Elemental activities, such as catalysis or binding, describing the actions of a gene product at the molecular level. A given gene product may exhibit one or more molecular functions.
signal transducer activity
Mediates the transfer of a signal from the outside to the inside of a cell by means other than the introduction of the signal molecule itself into the cell.
receptor activity
Combining with an extracellular or intracellular messenger to initiate a change in cell activity.
transmembrane receptor activity
Combining with an extracellular or intracellular messenger to initiate a change in cell activity, and spanning to the membrane of either the cell or an organelle.
G-protein coupled receptor activity
A receptor that binds an extracellular ligand and transmits the signal to a heterotrimeric G-protein complex. These receptors are characteristically seven-transmembrane receptors and are made up of hetero- or homodimers.
molecular transducer activity
The molecular function that accepts an input of one form and creates an output of a different form.
all
This term is the most general term possible
| Id | Pvalue | ExpCount | Count | Size | Term |
|---|---|---|---|---|---|
| 04080 | 1.367e-02 | 3.569 | 12 | 78 | Neuroactive ligand-receptor interaction |
| 00100 | 2.780e-02 | 1.052 | 6 | 23 | Biosynthesis of steroids |
ABHD5abhydrolase domain containing 5 (218739_at), score: -0.57 ACSF2acyl-CoA synthetase family member 2 (218844_at), score: 0.54 ACSL4acyl-CoA synthetase long-chain family member 4 (202422_s_at), score: -0.62 ACTC1actin, alpha, cardiac muscle 1 (205132_at), score: -0.65 ACTR3BARP3 actin-related protein 3 homolog B (yeast) (218868_at), score: -0.73 ADARadenosine deaminase, RNA-specific (201786_s_at), score: 0.64 ADH1Balcohol dehydrogenase 1B (class I), beta polypeptide (209612_s_at), score: 0.76 ADORA2Badenosine A2b receptor (205891_at), score: 0.77 AHI1Abelson helper integration site 1 (221569_at), score: -0.8 AIM1absent in melanoma 1 (212543_at), score: 0.71 AKR1C3aldo-keto reductase family 1, member C3 (3-alpha hydroxysteroid dehydrogenase, type II) (209160_at), score: 0.64 ALDH3A2aldehyde dehydrogenase 3 family, member A2 (202053_s_at), score: 0.56 ANGPTL2angiopoietin-like 2 (213004_at), score: 0.76 ANK2ankyrin 2, neuronal (202920_at), score: 0.64 ANXA10annexin A10 (210143_at), score: -0.91 APOBEC3Gapolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 3G (204205_at), score: 0.55 APOL3apolipoprotein L, 3 (221087_s_at), score: 0.55 APOL6apolipoprotein L, 6 (219716_at), score: 0.85 ARHGAP6Rho GTPase activating protein 6 (206167_s_at), score: 0.58 ARHGEF2rho/rac guanine nucleotide exchange factor (GEF) 2 (209435_s_at), score: 0.81 ARHGEF3Rho guanine nucleotide exchange factor (GEF) 3 (218501_at), score: 0.53 ARL4CADP-ribosylation factor-like 4C (202207_at), score: 0.73 ARNTL2aryl hydrocarbon receptor nuclear translocator-like 2 (220658_s_at), score: -0.64 ASAP3ArfGAP with SH3 domain, ankyrin repeat and PH domain 3 (219103_at), score: 0.65 ATF4activating transcription factor 4 (tax-responsive enhancer element B67) (200779_at), score: 0.58 ATP6V0BATPase, H+ transporting, lysosomal 21kDa, V0 subunit b (200078_s_at), score: -0.67 B3GALT4UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 4 (210205_at), score: 0.55 BBC3BCL2 binding component 3 (211692_s_at), score: 0.62 BCAS3breast carcinoma amplified sequence 3 (220488_s_at), score: 0.66 BDKRB2bradykinin receptor B2 (205870_at), score: 0.71 BEX4brain expressed, X-linked 4 (215440_s_at), score: 0.56 BHLHE40basic helix-loop-helix family, member e40 (201170_s_at), score: 0.59 BIN1bridging integrator 1 (210201_x_at), score: 0.67 BMP6bone morphogenetic protein 6 (206176_at), score: -0.66 BNC2basonuclin 2 (220272_at), score: 0.57 BRIP1BRCA1 interacting protein C-terminal helicase 1 (221703_at), score: -0.61 BTG2BTG family, member 2 (201236_s_at), score: 0.85 C11orf41chromosome 11 open reading frame 41 (214772_at), score: -0.63 C13orf15chromosome 13 open reading frame 15 (218723_s_at), score: 0.93 C14orf159chromosome 14 open reading frame 159 (218298_s_at), score: 0.64 C1orf107chromosome 1 open reading frame 107 (214193_s_at), score: -0.58 C1orf38chromosome 1 open reading frame 38 (210785_s_at), score: 0.65 C4orf18chromosome 4 open reading frame 18 (219872_at), score: 0.6 C4orf43chromosome 4 open reading frame 43 (218513_at), score: -0.61 C5orf23chromosome 5 open reading frame 23 (219054_at), score: -0.55 C5orf4chromosome 5 open reading frame 4 (48031_r_at), score: 0.55 C6orf48chromosome 6 open reading frame 48 (220755_s_at), score: 0.54 C7orf64chromosome 7 open reading frame 64 (221596_s_at), score: -0.58 C9orf91chromosome 9 open reading frame 91 (221865_at), score: 0.54 CALCOCO1calcium binding and coiled-coil domain 1 (209002_s_at), score: 0.78 CAMLGcalcium modulating ligand (203538_at), score: 0.55 CBFBcore-binding factor, beta subunit (206788_s_at), score: -0.55 CBScystathionine-beta-synthase (212816_s_at), score: 0.61 CCDC28Acoiled-coil domain containing 28A (209479_at), score: 0.76 CCL11chemokine (C-C motif) ligand 11 (210133_at), score: 0.64 CCNE2cyclin E2 (205034_at), score: -0.58 CCNG1cyclin G1 (208796_s_at), score: 0.61 CCNT2cyclin T2 (204645_at), score: -0.58 CDC42EP2CDC42 effector protein (Rho GTPase binding) 2 (209850_s_at), score: 0.67 CDCP1CUB domain containing protein 1 (218451_at), score: -0.65 CDH6cadherin 6, type 2, K-cadherin (fetal kidney) (205532_s_at), score: -0.68 CEBPGCCAAT/enhancer binding protein (C/EBP), gamma (204203_at), score: 0.57 CEP68centrosomal protein 68kDa (212677_s_at), score: 0.56 CHORDC1cysteine and histidine-rich domain (CHORD)-containing 1 (218566_s_at), score: -0.64 CHUKconserved helix-loop-helix ubiquitous kinase (209666_s_at), score: -0.68 CLASP2cytoplasmic linker associated protein 2 (212306_at), score: -0.64 CLDN11claudin 11 (206908_s_at), score: 0.61 CLIP3CAP-GLY domain containing linker protein 3 (212358_at), score: 0.68 COL21A1collagen, type XXI, alpha 1 (208096_s_at), score: 0.55 CRBNcereblon (218142_s_at), score: 0.75 CRTC3CREB regulated transcription coactivator 3 (218648_at), score: 0.88 CTDSP2CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 2 (203445_s_at), score: 0.62 CXCL10chemokine (C-X-C motif) ligand 10 (204533_at), score: 0.55 DAAM2dishevelled associated activator of morphogenesis 2 (212793_at), score: 0.78 DAPK1death-associated protein kinase 1 (203139_at), score: 0.88 DCKdeoxycytidine kinase (203302_at), score: -0.64 DCLK1doublecortin-like kinase 1 (205399_at), score: 0.8 DDAH2dimethylarginine dimethylaminohydrolase 2 (215537_x_at), score: 0.66 DDIT3DNA-damage-inducible transcript 3 (209383_at), score: 0.55 DDX58DEAD (Asp-Glu-Ala-Asp) box polypeptide 58 (218943_s_at), score: 0.64 DDX60DEAD (Asp-Glu-Ala-Asp) box polypeptide 60 (218986_s_at), score: 0.54 DENND2ADENN/MADD domain containing 2A (53991_at), score: 0.75 DEPDC6DEP domain containing 6 (218858_at), score: 0.9 DGKIdiacylglycerol kinase, iota (206806_at), score: -0.63 DHRS3dehydrogenase/reductase (SDR family) member 3 (202481_at), score: 0.85 DIRAS3DIRAS family, GTP-binding RAS-like 3 (215506_s_at), score: -0.77 DLX2distal-less homeobox 2 (207147_at), score: -0.69 DNAJB9DnaJ (Hsp40) homolog, subfamily B, member 9 (202843_at), score: -0.62 DOCK10dedicator of cytokinesis 10 (219279_at), score: -0.86 DPH5DPH5 homolog (S. cerevisiae) (219590_x_at), score: 0.54 DRAMdamage-regulated autophagy modulator (218627_at), score: 0.65 DTX4deltex homolog 4 (Drosophila) (212611_at), score: 0.6 DUSP4dual specificity phosphatase 4 (204014_at), score: -0.68 EAF2ELL associated factor 2 (219551_at), score: -0.63 EIF2B5eukaryotic translation initiation factor 2B, subunit 5 epsilon, 82kDa (212351_at), score: -0.55 EIF4Eeukaryotic translation initiation factor 4E (201436_at), score: -0.8 ELTD1EGF, latrophilin and seven transmembrane domain containing 1 (219134_at), score: -0.57 ENOX1ecto-NOX disulfide-thiol exchanger 1 (219501_at), score: 0.85 EPB41L4Berythrocyte membrane protein band 4.1 like 4B (220161_s_at), score: -0.55 EPHB6EPH receptor B6 (204718_at), score: 0.64 EPS8L2EPS8-like 2 (218180_s_at), score: 0.59 ESM1endothelial cell-specific molecule 1 (208394_x_at), score: -0.72 FAM102Afamily with sequence similarity 102, member A (212400_at), score: 0.6 FAM110Bfamily with sequence similarity 110, member B (221959_at), score: 0.54 FARSAphenylalanyl-tRNA synthetase, alpha subunit (216602_s_at), score: -0.56 FBXL7F-box and leucine-rich repeat protein 7 (213249_at), score: 0.56 FERMT1fermitin family homolog 1 (Drosophila) (218796_at), score: -0.66 FGF9fibroblast growth factor 9 (glia-activating factor) (206404_at), score: 0.86 FGGYFGGY carbohydrate kinase domain containing (219718_at), score: 0.56 FICDFIC domain containing (219910_at), score: -0.55 FKBP9FK506 binding protein 9, 63 kDa (212169_at), score: 0.54 FLT3LGfms-related tyrosine kinase 3 ligand (210607_at), score: 0.56 FNBP1formin binding protein 1 (212288_at), score: 0.67 FYNFYN oncogene related to SRC, FGR, YES (210105_s_at), score: 0.55 G0S2G0/G1switch 2 (213524_s_at), score: -0.59 GABBR2gamma-aminobutyric acid (GABA) B receptor, 2 (209990_s_at), score: -0.58 GALgalanin prepropeptide (214240_at), score: -0.67 GAL3ST4galactose-3-O-sulfotransferase 4 (219815_at), score: 0.55 GALK2galactokinase 2 (205219_s_at), score: -0.64 GALNAC4S-6STB cell RAG associated protein (203066_at), score: 0.65 GALNT12UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 12 (GalNAc-T12) (218885_s_at), score: 0.71 GAS7growth arrest-specific 7 (202191_s_at), score: 0.57 GDF15growth differentiation factor 15 (221577_x_at), score: 0.78 GDF5growth differentiation factor 5 (206614_at), score: 0.56 GDPD5glycerophosphodiester phosphodiesterase domain containing 5 (32502_at), score: 0.85 GFOD1glucose-fructose oxidoreductase domain containing 1 (219821_s_at), score: -0.68 GKglycerol kinase (207387_s_at), score: -0.81 GK3Pglycerol kinase 3 pseudogene (215966_x_at), score: -0.85 GLI3GLI family zinc finger 3 (205201_at), score: 0.58 GLTSCR2glioma tumor suppressor candidate region gene 2 (217807_s_at), score: 0.68 GMCL1germ cell-less homolog 1 (Drosophila) (218458_at), score: -0.68 GMPRguanosine monophosphate reductase (204187_at), score: 0.55 GOLT1Bgolgi transport 1 homolog B (S. cerevisiae) (218193_s_at), score: -0.62 GPD2glycerol-3-phosphate dehydrogenase 2 (mitochondrial) (210007_s_at), score: -0.59 GPRC5BG protein-coupled receptor, family C, group 5, member B (203632_s_at), score: 0.64 GRAMD1BGRAM domain containing 1B (212906_at), score: -0.58 GRB14growth factor receptor-bound protein 14 (206204_at), score: -0.74 GTF2A1Lgeneral transcription factor IIA, 1-like (213413_at), score: 1 GUCY1B3guanylate cyclase 1, soluble, beta 3 (203817_at), score: 0.67 GUF1GUF1 GTPase homolog (S. cerevisiae) (218884_s_at), score: -0.57 GYPCglycophorin C (Gerbich blood group) (202947_s_at), score: 0.57 HERC6hect domain and RLD 6 (219352_at), score: 0.63 HHLA3HERV-H LTR-associating 3 (220387_s_at), score: 0.54 HISPPD1histidine acid phosphatase domain containing 1 (203253_s_at), score: -0.67 HIST1H2BChistone cluster 1, H2bc (214455_at), score: -0.55 HIST3H2Ahistone cluster 3, H2a (221582_at), score: 0.58 HMGCR3-hydroxy-3-methylglutaryl-Coenzyme A reductase (202539_s_at), score: 0.65 HMGCS13-hydroxy-3-methylglutaryl-Coenzyme A synthase 1 (soluble) (221750_at), score: 0.54 HOXC5homeobox C5 (206739_at), score: -0.58 HRH1histamine receptor H1 (205580_s_at), score: 0.57 HSD17B7hydroxysteroid (17-beta) dehydrogenase 7 (220081_x_at), score: 0.83 HSPA12Aheat shock 70kDa protein 12A (214434_at), score: 0.7 HTR2A5-hydroxytryptamine (serotonin) receptor 2A (207135_at), score: -0.55 ID4inhibitor of DNA binding 4, dominant negative helix-loop-helix protein (209291_at), score: -0.69 IDI1isopentenyl-diphosphate delta isomerase 1 (204615_x_at), score: 0.64 IFI35interferon-induced protein 35 (209417_s_at), score: 0.58 IFI44Linterferon-induced protein 44-like (204439_at), score: 0.57 IFIH1interferon induced with helicase C domain 1 (219209_at), score: 0.93 IFIT3interferon-induced protein with tetratricopeptide repeats 3 (204747_at), score: 0.68 IGBP1immunoglobulin (CD79A) binding protein 1 (202105_at), score: 0.71 IL13RA2interleukin 13 receptor, alpha 2 (206172_at), score: -0.75 IL33interleukin 33 (209821_at), score: -0.8 IL7Rinterleukin 7 receptor (205798_at), score: -0.65 INPP5Binositol polyphosphate-5-phosphatase, 75kDa (213804_at), score: 0.68 INSIG1insulin induced gene 1 (201627_s_at), score: 0.61 IRF1interferon regulatory factor 1 (202531_at), score: 0.69 IRF7interferon regulatory factor 7 (208436_s_at), score: 0.58 IRF9interferon regulatory factor 9 (203882_at), score: 0.55 ITGA2integrin, alpha 2 (CD49B, alpha 2 subunit of VLA-2 receptor) (205032_at), score: -0.58 JUNDjun D proto-oncogene (203751_x_at), score: 0.66 KAL1Kallmann syndrome 1 sequence (205206_at), score: -0.66 KCNB1potassium voltage-gated channel, Shab-related subfamily, member 1 (211006_s_at), score: 0.68 KCND3potassium voltage-gated channel, Shal-related subfamily, member 3 (213832_at), score: 0.61 KCNJ8potassium inwardly-rectifying channel, subfamily J, member 8 (205303_at), score: -0.66 KIAA0232KIAA0232 (212441_at), score: 0.56 KIAA1305KIAA1305 (220911_s_at), score: 0.67 KIAA1462KIAA1462 (213316_at), score: 0.77 KLHDC2kelch domain containing 2 (217906_at), score: 0.56 KLHL23kelch-like 23 (Drosophila) (213610_s_at), score: -0.65 KLHL7kelch-like 7 (Drosophila) (220239_at), score: -0.58 LARP4La ribonucleoprotein domain family, member 4 (214155_s_at), score: -0.67 LDLRlow density lipoprotein receptor (202068_s_at), score: 0.78 LEPREL1leprecan-like 1 (218717_s_at), score: -0.54 LETMD1LETM1 domain containing 1 (207170_s_at), score: 0.61 LIMK2LIM domain kinase 2 (202193_at), score: -0.58 LIN7Blin-7 homolog B (C. elegans) (219760_at), score: -0.64 LMCD1LIM and cysteine-rich domains 1 (218574_s_at), score: 0.55 LOC100132540similar to LOC339047 protein (214870_x_at), score: 0.86 LOC339047hypothetical protein LOC339047 (221501_x_at), score: 0.85 LOC399491LOC399491 protein (214035_x_at), score: 0.81 LPIN1lipin 1 (212274_at), score: 0.62 LRRN3leucine rich repeat neuronal 3 (209841_s_at), score: -0.59 LSSlanosterol synthase (2,3-oxidosqualene-lanosterol cyclase) (211019_s_at), score: 0.82 LYNv-yes-1 Yamaguchi sarcoma viral related oncogene homolog (202625_at), score: -0.58 LYPD1LY6/PLAUR domain containing 1 (212909_at), score: -0.75 LYRM1LYR motif containing 1 (203897_at), score: 0.55 MAFv-maf musculoaponeurotic fibrosarcoma oncogene homolog (avian) (209348_s_at), score: 0.64 MAN1C1mannosidase, alpha, class 1C, member 1 (218918_at), score: 0.81 MAP2K5mitogen-activated protein kinase kinase 5 (211370_s_at), score: 0.61 MAP2K6mitogen-activated protein kinase kinase 6 (205698_s_at), score: 0.54 MARCH3membrane-associated ring finger (C3HC4) 3 (213256_at), score: -0.63 MBPmyelin basic protein (210136_at), score: 0.81 MC4Rmelanocortin 4 receptor (221467_at), score: -0.76 MEF2Cmyocyte enhancer factor 2C (209199_s_at), score: -0.55 MFAP3Lmicrofibrillar-associated protein 3-like (205442_at), score: -0.67 MGC87042similar to Six transmembrane epithelial antigen of prostate (217553_at), score: -0.75 MIS12MIS12, MIND kinetochore complex component, homolog (yeast) (221559_s_at), score: -0.57 MKNK2MAP kinase interacting serine/threonine kinase 2 (218205_s_at), score: 0.73 MMP10matrix metallopeptidase 10 (stromelysin 2) (205680_at), score: -0.75 MMP16matrix metallopeptidase 16 (membrane-inserted) (207012_at), score: -0.81 MMP3matrix metallopeptidase 3 (stromelysin 1, progelatinase) (205828_at), score: -0.56 MOCOSmolybdenum cofactor sulfurase (219959_at), score: 0.66 MSTO1misato homolog 1 (Drosophila) (218296_x_at), score: -0.78 MT1Xmetallothionein 1X (204326_x_at), score: -0.56 MTAPmethylthioadenosine phosphorylase (211363_s_at), score: -0.56 MTUS1mitochondrial tumor suppressor 1 (212096_s_at), score: 0.55 MVDmevalonate (diphospho) decarboxylase (203027_s_at), score: 0.55 MX1myxovirus (influenza virus) resistance 1, interferon-inducible protein p78 (mouse) (202086_at), score: 0.66 MX2myxovirus (influenza virus) resistance 2 (mouse) (204994_at), score: 0.83 MYBBP1AMYB binding protein (P160) 1a (219098_at), score: -0.6 NCALDneurocalcin delta (211685_s_at), score: -0.65 NEDD4neural precursor cell expressed, developmentally down-regulated 4 (213012_at), score: -0.61 NEFMneurofilament, medium polypeptide (205113_at), score: -0.86 NETO2neuropilin (NRP) and tolloid (TLL)-like 2 (218888_s_at), score: -0.69 NINJ1ninjurin 1 (203045_at), score: 0.55 NMIN-myc (and STAT) interactor (203964_at), score: 0.63 NOX4NADPH oxidase 4 (219773_at), score: -0.6 NPnucleoside phosphorylase (201695_s_at), score: -0.56 NPIPnuclear pore complex interacting protein (204538_x_at), score: 0.87 NPR3natriuretic peptide receptor C/guanylate cyclase C (atrionatriuretic peptide receptor C) (219789_at), score: -0.58 NPTX1neuronal pentraxin I (204684_at), score: -0.88 NR1H3nuclear receptor subfamily 1, group H, member 3 (203920_at), score: 0.58 NR5A2nuclear receptor subfamily 5, group A, member 2 (208343_s_at), score: -0.57 NTMneurotrimin (222020_s_at), score: -0.64 NUP160nucleoporin 160kDa (212709_at), score: -0.62 OAS12',5'-oligoadenylate synthetase 1, 40/46kDa (205552_s_at), score: 0.65 OAS22'-5'-oligoadenylate synthetase 2, 69/71kDa (204972_at), score: 0.75 OBFC2Aoligonucleotide/oligosaccharide-binding fold containing 2A (219334_s_at), score: -0.59 OPCMLopioid binding protein/cell adhesion molecule-like (214111_at), score: -0.66 ORAI3ORAI calcium release-activated calcium modulator 3 (221864_at), score: 0.89 OSR2odd-skipped related 2 (Drosophila) (213568_at), score: 0.55 OSTM1osteopetrosis associated transmembrane protein 1 (218196_at), score: -0.6 P2RY5purinergic receptor P2Y, G-protein coupled, 5 (218589_at), score: 0.64 PAQR3progestin and adipoQ receptor family member III (213372_at), score: -0.69 PCDH9protocadherin 9 (219737_s_at), score: -0.8 PCK2phosphoenolpyruvate carboxykinase 2 (mitochondrial) (202847_at), score: 0.58 PCYT2phosphate cytidylyltransferase 2, ethanolamine (209577_at), score: 0.7 PDCD1LG2programmed cell death 1 ligand 2 (220049_s_at), score: -0.6 PDE4Aphosphodiesterase 4A, cAMP-specific (phosphodiesterase E2 dunce homolog, Drosophila) (204735_at), score: 0.54 PDE5Aphosphodiesterase 5A, cGMP-specific (206757_at), score: 0.6 PDGFRBplatelet-derived growth factor receptor, beta polypeptide (202273_at), score: 0.62 PDK2pyruvate dehydrogenase kinase, isozyme 2 (202590_s_at), score: 0.62 PDSS1prenyl (decaprenyl) diphosphate synthase, subunit 1 (220865_s_at), score: -0.61 PIRpirin (iron-binding nuclear protein) (207469_s_at), score: 0.6 PITX1paired-like homeodomain 1 (208502_s_at), score: 0.58 PKIAprotein kinase (cAMP-dependent, catalytic) inhibitor alpha (204612_at), score: -0.58 PKIGprotein kinase (cAMP-dependent, catalytic) inhibitor gamma (202732_at), score: 0.66 PLA2G4Cphospholipase A2, group IVC (cytosolic, calcium-independent) (209785_s_at), score: 0.56 PLEKHA5pleckstrin homology domain containing, family A member 5 (220952_s_at), score: 0.62 PLEKHO2pleckstrin homology domain containing, family O member 2 (204436_at), score: 0.61 PMP22peripheral myelin protein 22 (210139_s_at), score: 0.76 PNPLA2patatin-like phospholipase domain containing 2 (212705_x_at), score: 0.56 POLR3Gpolymerase (RNA) III (DNA directed) polypeptide G (32kD) (206653_at), score: -0.6 PPARGperoxisome proliferator-activated receptor gamma (208510_s_at), score: 0.6 PPATphosphoribosyl pyrophosphate amidotransferase (209433_s_at), score: -0.78 PPLperiplakin (203407_at), score: 0.85 PPP3R1protein phosphatase 3 (formerly 2B), regulatory subunit B, alpha isoform (204507_s_at), score: -0.67 PRELPproline/arginine-rich end leucine-rich repeat protein (204223_at), score: 0.65 PRPS2phosphoribosyl pyrophosphate synthetase 2 (203401_at), score: -0.66 PRR5proline rich 5 (renal) (47069_at), score: 0.61 PRSS2protease, serine, 2 (trypsin 2) (205402_x_at), score: -0.65 PRSS3protease, serine, 3 (213421_x_at), score: -0.61 PTGER4prostaglandin E receptor 4 (subtype EP4) (204897_at), score: 0.64 PTGS1prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) (215813_s_at), score: -0.63 RAB7ARAB7A, member RAS oncogene family (211960_s_at), score: -0.61 RAGErenal tumor antigen (205130_at), score: -0.58 RANBP6RAN binding protein 6 (213019_at), score: -0.61 RASSF2Ras association (RalGDS/AF-6) domain family member 2 (203185_at), score: 0.77 RCAN2regulator of calcineurin 2 (203498_at), score: 0.54 REREarginine-glutamic acid dipeptide (RE) repeats (200940_s_at), score: 0.59 RGS20regulator of G-protein signaling 20 (210138_at), score: -0.55 RHBDF1rhomboid 5 homolog 1 (Drosophila) (218686_s_at), score: 0.54 RINT1RAD50 interactor 1 (218598_at), score: -0.55 RNF187ring finger protein 187 (212155_at), score: 0.54 RNF44ring finger protein 44 (203286_at), score: 0.57 ROR1receptor tyrosine kinase-like orphan receptor 1 (205805_s_at), score: 0.69 ROR2receptor tyrosine kinase-like orphan receptor 2 (205578_at), score: 0.73 RP11-345P4.4similar to solute carrier family 35, member E2 (217122_s_at), score: 0.58 RPS6KA6ribosomal protein S6 kinase, 90kDa, polypeptide 6 (220738_s_at), score: -0.59 RSC1A1regulatory solute carrier protein, family 1, member 1 (214583_at), score: -0.65 S1PR1sphingosine-1-phosphate receptor 1 (204642_at), score: -0.62 SAV1salvador homolog 1 (Drosophila) (218276_s_at), score: 0.65 SDF2L1stromal cell-derived factor 2-like 1 (218681_s_at), score: -0.61 SECTM1secreted and transmembrane 1 (213716_s_at), score: 0.6 SELENBP1selenium binding protein 1 (214433_s_at), score: 0.55 SELPLGselectin P ligand (209879_at), score: -0.63 SERPINB2serpin peptidase inhibitor, clade B (ovalbumin), member 2 (204614_at), score: -0.72 SESN1sestrin 1 (218346_s_at), score: 0.65 SIGIRRsingle immunoglobulin and toll-interleukin 1 receptor (TIR) domain (52940_at), score: 0.57 SIX2SIX homeobox 2 (206510_at), score: 0.84 SLC25A13solute carrier family 25, member 13 (citrin) (203775_at), score: -0.74 SLC27A3solute carrier family 27 (fatty acid transporter), member 3 (222217_s_at), score: 0.62 SLC2A6solute carrier family 2 (facilitated glucose transporter), member 6 (220091_at), score: 0.67 SLC4A7solute carrier family 4, sodium bicarbonate cotransporter, member 7 (209884_s_at), score: -0.79 SLC6A9solute carrier family 6 (neurotransmitter transporter, glycine), member 9 (207043_s_at), score: 0.76 SLMO2slowmo homolog 2 (Drosophila) (217851_s_at), score: -0.63 SMAD3SMAD family member 3 (218284_at), score: 0.57 SMN1survival of motor neuron 1, telomeric (203852_s_at), score: -0.61 SNNstannin (218032_at), score: 0.66 SOD2superoxide dismutase 2, mitochondrial (221477_s_at), score: 0.53 SPRY1sprouty homolog 1, antagonist of FGF signaling (Drosophila) (212558_at), score: 0.66 SQLEsqualene epoxidase (213562_s_at), score: 0.6 SREBF1sterol regulatory element binding transcription factor 1 (202308_at), score: 0.7 SREBF2sterol regulatory element binding transcription factor 2 (201247_at), score: 0.62 SRGAP2SLIT-ROBO Rho GTPase activating protein 2 (213329_at), score: 0.6 SSTR1somatostatin receptor 1 (208482_at), score: -0.78 STARD5StAR-related lipid transfer (START) domain containing 5 (213820_s_at), score: 0.55 SV2Asynaptic vesicle glycoprotein 2A (203069_at), score: 0.73 SYNPOsynaptopodin (202796_at), score: 0.75 TAF13TAF13 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 18kDa (205966_at), score: -0.56 TAF1ATATA box binding protein (TBP)-associated factor, RNA polymerase I, A, 48kDa (206613_s_at), score: -0.71 TBC1D12TBC1 domain family, member 12 (221858_at), score: 0.63 TBC1D2BTBC1 domain family, member 2B (212796_s_at), score: 0.72 TBC1D8TBC1 domain family, member 8 (with GRAM domain) (204526_s_at), score: 0.86 TCEA2transcription elongation factor A (SII), 2 (203919_at), score: 0.61 TENC1tensin like C1 domain containing phosphatase (tensin 2) (212494_at), score: 0.65 TGDSTDP-glucose 4,6-dehydratase (208249_s_at), score: -0.58 THUMPD2THUMP domain containing 2 (219248_at), score: -0.55 TICAM2toll-like receptor adaptor molecule 2 (214658_at), score: -0.6 TLR3toll-like receptor 3 (206271_at), score: 0.74 TM4SF1transmembrane 4 L six family member 1 (215034_s_at), score: -0.82 TMEFF1transmembrane protein with EGF-like and two follistatin-like domains 1 (205122_at), score: -0.58 TMEM159transmembrane protein 159 (213272_s_at), score: 0.57 TMEM2transmembrane protein 2 (218113_at), score: -0.76 TMEM30Atransmembrane protein 30A (217743_s_at), score: -0.61 TNFAIP2tumor necrosis factor, alpha-induced protein 2 (202510_s_at), score: 0.59 TNFAIP6tumor necrosis factor, alpha-induced protein 6 (206026_s_at), score: 0.68 TNFSF9tumor necrosis factor (ligand) superfamily, member 9 (206907_at), score: 0.8 TNIP1TNFAIP3 interacting protein 1 (207196_s_at), score: 0.61 TNS3tensin 3 (217853_at), score: 0.7 TP53tumor protein p53 (201746_at), score: 0.73 TRIB3tribbles homolog 3 (Drosophila) (218145_at), score: 0.63 TRIM21tripartite motif-containing 21 (204804_at), score: 0.63 TRIM24tripartite motif-containing 24 (213301_x_at), score: -0.6 TRPC4transient receptor potential cation channel, subfamily C, member 4 (220817_at), score: -0.6 TSC22D3TSC22 domain family, member 3 (208763_s_at), score: 0.6 TSPAN13tetraspanin 13 (217979_at), score: -0.69 TUBGCP5tubulin, gamma complex associated protein 5 (214876_s_at), score: -0.63 UBE2L6ubiquitin-conjugating enzyme E2L 6 (201649_at), score: 0.61 UBXN1UBX domain protein 1 (201871_s_at), score: 0.55 UHRF1BP1LUHRF1 binding protein 1-like (213118_at), score: -0.7 ULBP2UL16 binding protein 2 (221291_at), score: -0.82 ULK1unc-51-like kinase 1 (C. elegans) (209333_at), score: 0.62 URB1URB1 ribosome biogenesis 1 homolog (S. cerevisiae) (212996_s_at), score: -0.68 VWA5Avon Willebrand factor A domain containing 5A (210102_at), score: 0.55 WBP2WW domain binding protein 2 (209117_at), score: 0.64 WDR19WD repeat domain 19 (220917_s_at), score: 0.62 WDR4WD repeat domain 4 (221632_s_at), score: -0.65 WNT2wingless-type MMTV integration site family member 2 (205648_at), score: 0.7 XAF1XIAP associated factor 1 (206133_at), score: 0.77 XYLT1xylosyltransferase I (213725_x_at), score: -0.57 YDD19YDD19 protein (37079_at), score: -0.58 YPEL5yippee-like 5 (Drosophila) (217783_s_at), score: 0.7 ZBTB20zinc finger and BTB domain containing 20 (205383_s_at), score: 0.55 ZBTB22zinc finger and BTB domain containing 22 (213081_at), score: 0.61 ZBTB6zinc finger and BTB domain containing 6 (206098_at), score: -0.59 ZC3H12Azinc finger CCCH-type containing 12A (218810_at), score: 0.66 ZDHHC7zinc finger, DHHC-type containing 7 (218606_at), score: 0.79 ZNF141zinc finger protein 141 (206931_at), score: -0.55 ZNF804Azinc finger protein 804A (215767_at), score: -0.64
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-TABM-263-raw-cel-1515486371.cel | 37 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485891.cel | 13 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485811.cel | 9 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485771.cel | 7 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486331.cel | 35 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486271.cel | 32 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486291.cel | 33 | 6 | Cycle | hgu133a2 | none | Cycle 1 |