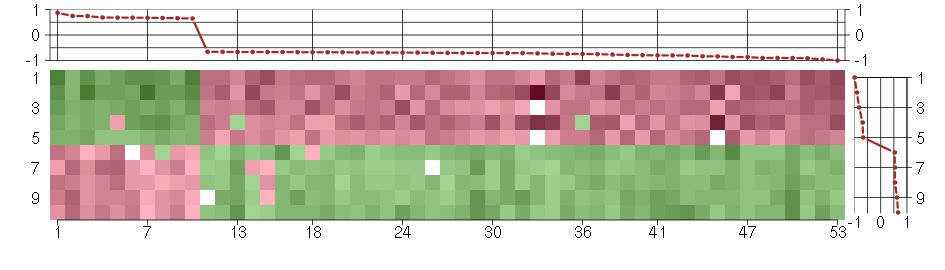

Under-expression is coded with green,
over-expression with red color.


extracellular region
The space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers the host cell environment outside an intracellular parasite.
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
extracellular space
That part of a multicellular organism outside the cells proper, usually taken to be outside the plasma membranes, and occupied by fluid.
extracellular region part
Any constituent part of the extracellular region, the space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers constituent parts of the host cell environment outside an intracellular parasite.
all
This term is the most general term possible
extracellular region part
Any constituent part of the extracellular region, the space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers constituent parts of the host cell environment outside an intracellular parasite.

protein binding
Interacting selectively with any protein or protein complex (a complex of two or more proteins that may include other nonprotein molecules).
molecular_function
Elemental activities, such as catalysis or binding, describing the actions of a gene product at the molecular level. A given gene product may exhibit one or more molecular functions.
receptor binding
Interacting selectively with one or more specific sites on a receptor molecule, a macromolecule that undergoes combination with a hormone, neurotransmitter, drug or intracellular messenger to initiate a change in cell function.
binding
The selective, often stoichiometric, interaction of a molecule with one or more specific sites on another molecule.
all
This term is the most general term possible
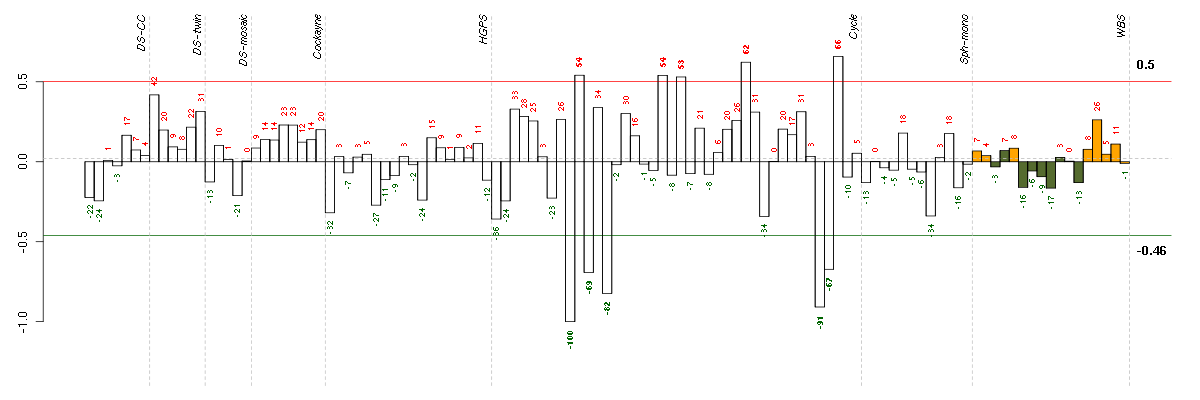
AHI1Abelson helper integration site 1 (221569_at), score: -0.78 AJAP1adherens junctions associated protein 1 (206460_at), score: -0.7 ANXA10annexin A10 (210143_at), score: -0.8 AREGamphiregulin (205239_at), score: -0.72 ARNTLaryl hydrocarbon receptor nuclear translocator-like (209824_s_at), score: -0.66 BMP6bone morphogenetic protein 6 (206176_at), score: -1 C21orf7chromosome 21 open reading frame 7 (221211_s_at), score: -0.67 C2CD2C2 calcium-dependent domain containing 2 (212875_s_at), score: -0.68 CBLBCas-Br-M (murine) ecotropic retroviral transforming sequence b (209682_at), score: -0.67 CD55CD55 molecule, decay accelerating factor for complement (Cromer blood group) (201925_s_at), score: -0.8 COL5A3collagen, type V, alpha 3 (52255_s_at), score: -0.71 CSGALNACT2chondroitin sulfate N-acetylgalactosaminyltransferase 2 (222235_s_at), score: -0.69 CXCL6chemokine (C-X-C motif) ligand 6 (granulocyte chemotactic protein 2) (206336_at), score: -0.74 DNAJB9DnaJ (Hsp40) homolog, subfamily B, member 9 (202843_at), score: -0.83 ENTPD7ectonucleoside triphosphate diphosphohydrolase 7 (220153_at), score: -0.73 ESM1endothelial cell-specific molecule 1 (208394_x_at), score: -0.8 FABP3fatty acid binding protein 3, muscle and heart (mammary-derived growth inhibitor) (214285_at), score: -0.68 FBXO31F-box protein 31 (219785_s_at), score: 0.66 FERMT1fermitin family homolog 1 (Drosophila) (218796_at), score: -0.9 FGF1fibroblast growth factor 1 (acidic) (205117_at), score: -0.69 FGF7fibroblast growth factor 7 (keratinocyte growth factor) (205782_at), score: -0.69 GKglycerol kinase (207387_s_at), score: -0.95 GK3Pglycerol kinase 3 pseudogene (215966_x_at), score: -0.91 GPR183G protein-coupled receptor 183 (205419_at), score: -0.84 GRB14growth factor receptor-bound protein 14 (206204_at), score: -0.79 HTR2A5-hydroxytryptamine (serotonin) receptor 2A (207135_at), score: -0.9 IL33interleukin 33 (209821_at), score: -0.91 KAL1Kallmann syndrome 1 sequence (205206_at), score: -0.86 KCNG1potassium voltage-gated channel, subfamily G, member 1 (214595_at), score: -0.77 MMP10matrix metallopeptidase 10 (stromelysin 2) (205680_at), score: -0.75 MREGmelanoregulin (219648_at), score: -0.71 MTMR9myotubularin related protein 9 (204837_at), score: -0.71 NCALDneurocalcin delta (211685_s_at), score: -0.74 NCAPD2non-SMC condensin I complex, subunit D2 (201774_s_at), score: 0.67 NEFLneurofilament, light polypeptide (221805_at), score: -0.67 PCDH9protocadherin 9 (219737_s_at), score: -0.68 PID1phosphotyrosine interaction domain containing 1 (219093_at), score: -0.68 PIONpigeon homolog (Drosophila) (222150_s_at), score: -0.69 PTHLHparathyroid hormone-like hormone (211756_at), score: -0.87 RFX5regulatory factor X, 5 (influences HLA class II expression) (202963_at), score: 0.74 RNF44ring finger protein 44 (203286_at), score: 0.68 SLC38A4solute carrier family 38, member 4 (220786_s_at), score: 0.68 SLC4A7solute carrier family 4, sodium bicarbonate cotransporter, member 7 (209884_s_at), score: -0.7 SMAD3SMAD family member 3 (218284_at), score: 0.65 TFDP2transcription factor Dp-2 (E2F dimerization partner 2) (203588_s_at), score: 0.87 TGDSTDP-glucose 4,6-dehydratase (208249_s_at), score: -0.67 TGFAtransforming growth factor, alpha (205016_at), score: -0.71 TMEM2transmembrane protein 2 (218113_at), score: -0.66 TMSB15Athymosin beta 15a (205347_s_at), score: 0.69 TRPC6transient receptor potential cation channel, subfamily C, member 6 (217287_s_at), score: -0.7 UCP2uncoupling protein 2 (mitochondrial, proton carrier) (208998_at), score: 0.75 URB1URB1 ribosome biogenesis 1 homolog (S. cerevisiae) (212996_s_at), score: -0.67 ZNF395zinc finger protein 395 (218149_s_at), score: 0.68
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-TABM-263-raw-cel-1515485811.cel | 9 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486351.cel | 36 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485891.cel | 13 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485851.cel | 11 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486371.cel | 37 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486051.cel | 21 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486011.cel | 19 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485831.cel | 10 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486191.cel | 28 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486391.cel | 38 | 6 | Cycle | hgu133a2 | none | Cycle 1 |