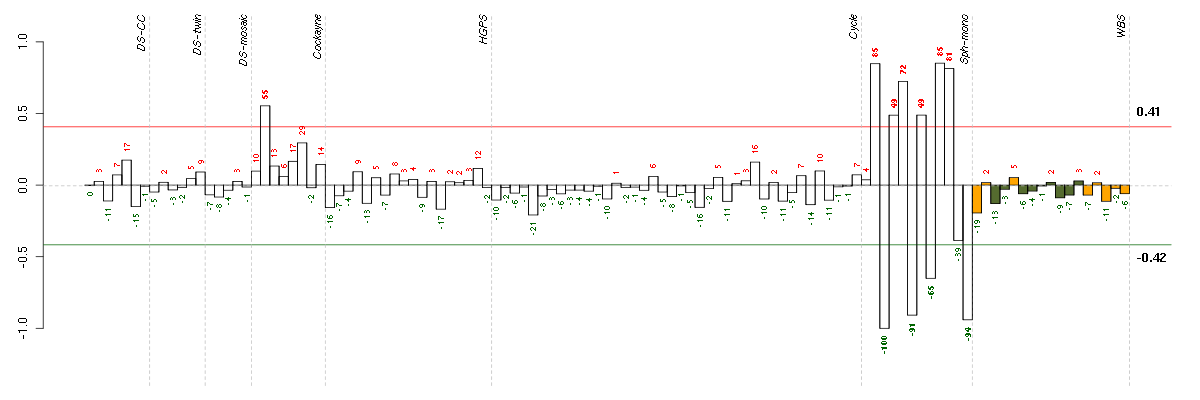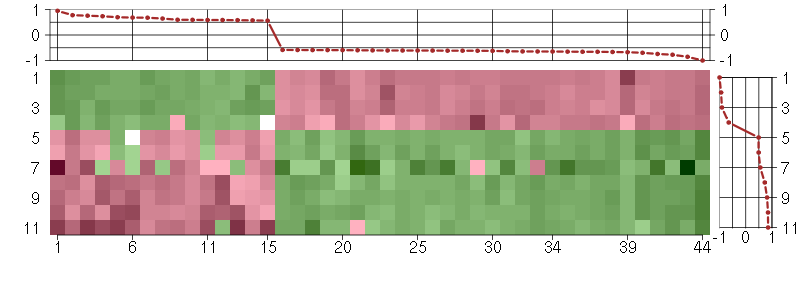

Under-expression is coded with green,
over-expression with red color.

multicellular organismal development
The biological process whose specific outcome is the progression of a multicellular organism over time from an initial condition (e.g. a zygote or a young adult) to a later condition (e.g. a multicellular animal or an aged adult).
anatomical structure morphogenesis
The process by which anatomical structures are generated and organized. Morphogenesis pertains to the creation of form.
organ morphogenesis
Morphogenesis of an organ. An organ is defined as a tissue or set of tissues that work together to perform a specific function or functions. Morphogenesis is the process by which anatomical structures are generated and organized. Organs are commonly observed as visibly distinct structures, but may also exist as loosely associated clusters of cells that work together to perform a specific function or functions.
nervous system development
The process whose specific outcome is the progression of nervous tissue over time, from its formation to its mature state.
biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
multicellular organismal process
Any biological process, occurring at the level of a multicellular organism, pertinent to its function.
developmental process
A biological process whose specific outcome is the progression of an integrated living unit: an anatomical structure (which may be a subcellular structure, cell, tissue, or organ), or organism over time from an initial condition to a later condition.
odontogenesis of dentine-containing tooth
The process whose specific outcome is the progression of a dentine-containing tooth over time, from its formation to the mature structure. A dentine-containing tooth is a hard, bony organ borne on the jaw or other bone of a vertebrate, and are composed mainly of dentine, a dense calcified substance, covered by a layer of enamel.
odontogenesis
The process whose specific outcome is the progression of a tooth or teeth over time, from formation to the mature structure(s). A tooth is any hard bony, calcareous, or chitinous organ found in the mouth or pharynx of an animal and used in procuring or masticating food.
organ development
Development of a tissue or tissues that work together to perform a specific function or functions. Development pertains to the process whose specific outcome is the progression of a structure over time, from its formation to the mature structure. Organs are commonly observed as visibly distinct structures, but may also exist as loosely associated clusters of cells that work together to perform a specific function or functions.
system development
The process whose specific outcome is the progression of an organismal system over time, from its formation to the mature structure. A system is a regularly interacting or interdependent group of organs or tissues that work together to carry out a given biological process.
anatomical structure development
The biological process whose specific outcome is the progression of an anatomical structure from an initial condition to its mature state. This process begins with the formation of the structure and ends with the mature structure, whatever form that may be including its natural destruction. An anatomical structure is any biological entity that occupies space and is distinguished from its surroundings. Anatomical structures can be macroscopic such as a carpel, or microscopic such as an acrosome.
all
This term is the most general term possible
multicellular organismal development
The biological process whose specific outcome is the progression of a multicellular organism over time from an initial condition (e.g. a zygote or a young adult) to a later condition (e.g. a multicellular animal or an aged adult).
anatomical structure morphogenesis
The process by which anatomical structures are generated and organized. Morphogenesis pertains to the creation of form.
system development
The process whose specific outcome is the progression of an organismal system over time, from its formation to the mature structure. A system is a regularly interacting or interdependent group of organs or tissues that work together to carry out a given biological process.
organ development
Development of a tissue or tissues that work together to perform a specific function or functions. Development pertains to the process whose specific outcome is the progression of a structure over time, from its formation to the mature structure. Organs are commonly observed as visibly distinct structures, but may also exist as loosely associated clusters of cells that work together to perform a specific function or functions.
organ morphogenesis
Morphogenesis of an organ. An organ is defined as a tissue or set of tissues that work together to perform a specific function or functions. Morphogenesis is the process by which anatomical structures are generated and organized. Organs are commonly observed as visibly distinct structures, but may also exist as loosely associated clusters of cells that work together to perform a specific function or functions.


AEBP1AE binding protein 1 (201792_at), score: -0.62 AKAP2A kinase (PRKA) anchor protein 2 (202759_s_at), score: -0.59 AKR1B10aldo-keto reductase family 1, member B10 (aldose reductase) (206561_s_at), score: 0.7 ARMCX1armadillo repeat containing, X-linked 1 (218694_at), score: -0.62 BMP7bone morphogenetic protein 7 (209590_at), score: 0.56 C14orf169chromosome 14 open reading frame 169 (219526_at), score: -0.65 CELSR1cadherin, EGF LAG seven-pass G-type receptor 1 (flamingo homolog, Drosophila) (41660_at), score: 0.59 CELSR3cadherin, EGF LAG seven-pass G-type receptor 3 (flamingo homolog, Drosophila) (40020_at), score: 0.78 COL3A1collagen, type III, alpha 1 (215076_s_at), score: -0.67 DGKDdiacylglycerol kinase, delta 130kDa (208072_s_at), score: 0.68 FAM174Bfamily with sequence similarity 174, member B (51158_at), score: -0.68 FCGR2AFc fragment of IgG, low affinity IIa, receptor (CD32) (203561_at), score: 0.58 FOXG1forkhead box G1 (206018_at), score: 0.59 GAL3ST4galactose-3-O-sulfotransferase 4 (219815_at), score: -0.59 GALCgalactosylceramidase (204417_at), score: -0.66 GREM1gremlin 1, cysteine knot superfamily, homolog (Xenopus laevis) (218469_at), score: -1 HSD17B12hydroxysteroid (17-beta) dehydrogenase 12 (217869_at), score: -0.61 IGSF3immunoglobulin superfamily, member 3 (202421_at), score: 0.74 IPWimprinted in Prader-Willi syndrome (non-protein coding) (221974_at), score: -0.62 ISL1ISL LIM homeobox 1 (206104_at), score: -0.59 KYNUkynureninase (L-kynurenine hydrolase) (217388_s_at), score: 0.95 LEF1lymphoid enhancer-binding factor 1 (221558_s_at), score: 0.76 LTBP2latent transforming growth factor beta binding protein 2 (204682_at), score: -0.59 LUMlumican (201744_s_at), score: -0.85 MPPED2metallophosphoesterase domain containing 2 (205413_at), score: 0.68 MYO1Bmyosin IB (212364_at), score: -0.77 NPTX1neuronal pentraxin I (204684_at), score: 0.6 PCLOpiccolo (presynaptic cytomatrix protein) (213558_at), score: 0.59 PDGFRAplatelet-derived growth factor receptor, alpha polypeptide (203131_at), score: -0.7 PEX5peroxisomal biogenesis factor 5 (203244_at), score: 0.57 PRR16proline rich 16 (220014_at), score: -0.6 PSTPIP2proline-serine-threonine phosphatase interacting protein 2 (219938_s_at), score: -0.64 PTPRDprotein tyrosine phosphatase, receptor type, D (214043_at), score: -0.62 RECKreversion-inducing-cysteine-rich protein with kazal motifs (205407_at), score: -0.6 RIPK4receptor-interacting serine-threonine kinase 4 (221215_s_at), score: 0.65 SULF1sulfatase 1 (212353_at), score: -0.61 SYNGR1synaptogyrin 1 (210613_s_at), score: -0.64 TBXA2Rthromboxane A2 receptor (336_at), score: -0.61 TCF7L2transcription factor 7-like 2 (T-cell specific, HMG-box) (212761_at), score: -0.59 TEStestis derived transcript (3 LIM domains) (202720_at), score: -0.74 THBS2thrombospondin 2 (203083_at), score: -0.66 TNS1tensin 1 (221748_s_at), score: -0.65 TSPYL5TSPY-like 5 (213122_at), score: -0.65 ULK2unc-51-like kinase 2 (C. elegans) (204062_s_at), score: -0.61
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-GEOD-4219-raw-cel-1311956178.cel | 6 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956824.cel | 24 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956358.cel | 10 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956418.cel | 13 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956275.cel | 8 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956398.cel | 12 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-3407-raw-cel-1437949579.cel | 2 | 4 | Cockayne | hgu133a | none | CSB |
| E-GEOD-4219-raw-cel-1311956321.cel | 9 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956614.cel | 18 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956138.cel | 4 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956457.cel | 14 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |