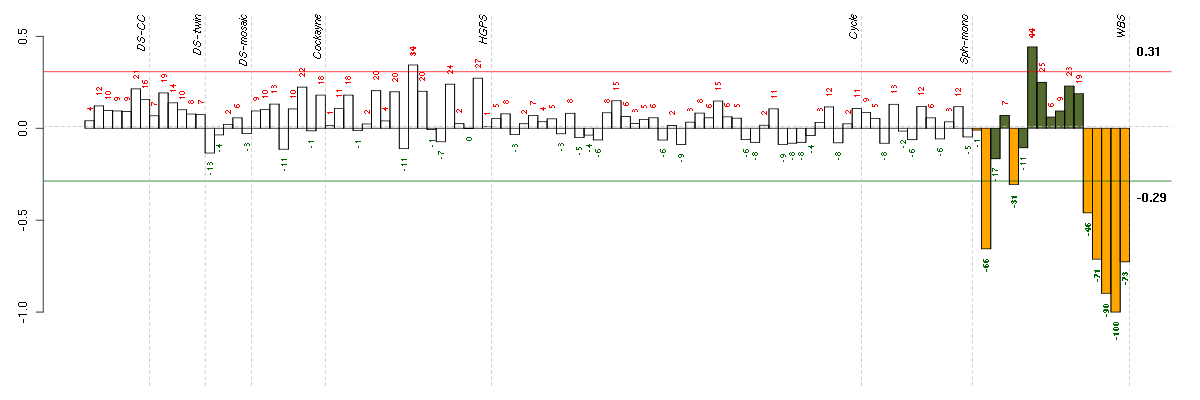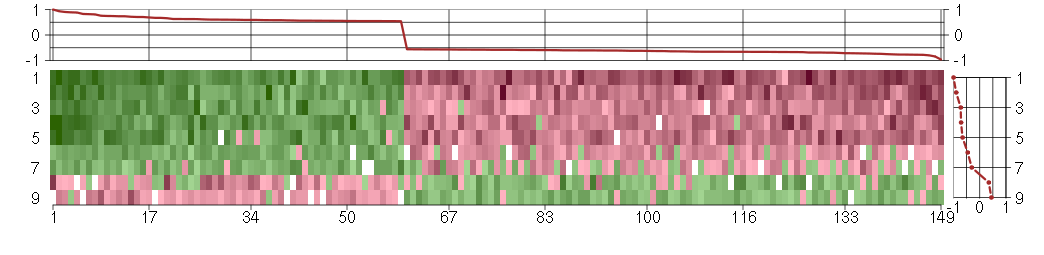

Under-expression is coded with green,
over-expression with red color.


membrane
Double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
integral to membrane
Penetrating at least one phospholipid bilayer of a membrane. May also refer to the state of being buried in the bilayer with no exposure outside the bilayer. When used to describe a protein, indicates that all or part of the peptide sequence is embedded in the membrane.
extracellular region
The space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers the host cell environment outside an intracellular parasite.
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
cell
The basic structural and functional unit of all organisms. Includes the plasma membrane and any external encapsulating structures such as the cell wall and cell envelope.
intrinsic to membrane
Located in a membrane such that some covalently attached portion of the gene product, for example part of a peptide sequence or some other covalently attached moiety such as a GPI anchor, spans or is embedded in one or both leaflets of the membrane.
membrane part
Any constituent part of a membrane, a double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
all
This term is the most general term possible
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
membrane part
Any constituent part of a membrane, a double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.

ABCC9ATP-binding cassette, sub-family C (CFTR/MRP), member 9 (208561_at), score: 0.55 ABHD14Aabhydrolase domain containing 14A (210006_at), score: -0.64 ACCN2amiloride-sensitive cation channel 2, neuronal (205156_s_at), score: -0.58 AGBL5ATP/GTP binding protein-like 5 (218480_at), score: -0.77 AKAP6A kinase (PRKA) anchor protein 6 (205359_at), score: -0.72 ALDH1A3aldehyde dehydrogenase 1 family, member A3 (203180_at), score: 0.71 B3GALT4UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 4 (210205_at), score: -0.6 B3GNT1UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 1 (203188_at), score: -0.71 BAZ1Bbromodomain adjacent to zinc finger domain, 1B (208445_s_at), score: 0.6 BCL7BB-cell CLL/lymphoma 7B (202518_at), score: 0.59 BST1bone marrow stromal cell antigen 1 (205715_at), score: -0.6 BTDbiotinidase (204167_at), score: -0.77 BTN3A1butyrophilin, subfamily 3, member A1 (209770_at), score: -0.6 BTN3A2butyrophilin, subfamily 3, member A2 (209846_s_at), score: -0.75 C10orf116chromosome 10 open reading frame 116 (203571_s_at), score: -0.66 C14orf94chromosome 14 open reading frame 94 (218383_at), score: -0.63 C15orf5chromosome 15 open reading frame 5 (208109_s_at), score: 0.73 C1orf115chromosome 1 open reading frame 115 (218546_at), score: 0.63 C4orf31chromosome 4 open reading frame 31 (219747_at), score: -0.59 C7orf58chromosome 7 open reading frame 58 (220032_at), score: -0.59 C9orf127chromosome 9 open reading frame 127 (207839_s_at), score: -0.96 CDH8cadherin 8, type 2 (217574_at), score: -0.77 CHST7carbohydrate (N-acetylglucosamine 6-O) sulfotransferase 7 (206756_at), score: -0.66 CLEC11AC-type lectin domain family 11, member A (211709_s_at), score: -0.59 CLEC3BC-type lectin domain family 3, member B (205200_at), score: -0.59 CLGNcalmegin (205830_at), score: 0.66 CNTNAP1contactin associated protein 1 (219400_at), score: -0.56 CRELD1cysteine-rich with EGF-like domains 1 (203368_at), score: -0.6 CRYBB2crystallin, beta B2 (206777_s_at), score: 0.54 CRYL1crystallin, lambda 1 (220753_s_at), score: -0.69 CST6cystatin E/M (206595_at), score: -0.57 CTPS2CTP synthase II (219080_s_at), score: -0.61 CYP26B1cytochrome P450, family 26, subfamily B, polypeptide 1 (219825_at), score: 0.67 DBC1deleted in bladder cancer 1 (205818_at), score: 0.83 DDIT3DNA-damage-inducible transcript 3 (209383_at), score: 0.62 DHRS1dehydrogenase/reductase (SDR family) member 1 (213279_at), score: -0.62 DKK3dickkopf homolog 3 (Xenopus laevis) (202196_s_at), score: -0.61 DNAJC4DnaJ (Hsp40) homolog, subfamily C, member 4 (206782_s_at), score: -0.66 EGFL6EGF-like-domain, multiple 6 (219454_at), score: 0.9 EIF4Heukaryotic translation initiation factor 4H (206621_s_at), score: 0.6 ELTD1EGF, latrophilin and seven transmembrane domain containing 1 (219134_at), score: 0.6 ENDOD1endonuclease domain containing 1 (212573_at), score: -0.69 F10coagulation factor X (205620_at), score: -0.69 FAM13Bfamily with sequence similarity 13, member B (218518_at), score: 0.73 FAT4FAT tumor suppressor homolog 4 (Drosophila) (219427_at), score: 0.61 FGD6FYVE, RhoGEF and PH domain containing 6 (219901_at), score: -0.57 FGF9fibroblast growth factor 9 (glia-activating factor) (206404_at), score: 0.93 FLGfilaggrin (215704_at), score: 0.72 GABREgamma-aminobutyric acid (GABA) A receptor, epsilon (204537_s_at), score: 0.67 GAS7growth arrest-specific 7 (202191_s_at), score: 0.56 GBP2guanylate binding protein 2, interferon-inducible (202748_at), score: -0.66 GCNT1glucosaminyl (N-acetyl) transferase 1, core 2 (beta-1,6-N-acetylglucosaminyltransferase) (205505_at), score: -0.62 GDPD5glycerophosphodiester phosphodiesterase domain containing 5 (32502_at), score: -0.63 GLRBglycine receptor, beta (205280_at), score: -0.62 GOLSYNGolgi-localized protein (218692_at), score: 0.88 GOT1glutamic-oxaloacetic transaminase 1, soluble (aspartate aminotransferase 1) (208813_at), score: 0.56 GSTM4glutathione S-transferase mu 4 (204149_s_at), score: -0.56 GSTT2glutathione S-transferase theta 2 (205439_at), score: -0.66 GTPBP2GTP binding protein 2 (221050_s_at), score: 0.63 HHLA3HERV-H LTR-associating 3 (220387_s_at), score: -0.58 HOXA7homeobox A7 (206848_at), score: 0.71 HSPA9heat shock 70kDa protein 9 (mortalin) (200690_at), score: 0.55 HSPB3heat shock 27kDa protein 3 (206375_s_at), score: -0.72 ID1inhibitor of DNA binding 1, dominant negative helix-loop-helix protein (208937_s_at), score: 0.69 IGBP1immunoglobulin (CD79A) binding protein 1 (202105_at), score: -0.58 IGF1insulin-like growth factor 1 (somatomedin C) (209541_at), score: 0.82 IGFBP7insulin-like growth factor binding protein 7 (201162_at), score: -0.6 IL1RL1interleukin 1 receptor-like 1 (207526_s_at), score: 0.55 IL27RAinterleukin 27 receptor, alpha (205926_at), score: -0.8 IL7interleukin 7 (206693_at), score: 0.56 ITGA7integrin, alpha 7 (216331_at), score: -0.57 ITGB8integrin, beta 8 (211488_s_at), score: 0.56 ITIH5inter-alpha (globulin) inhibitor H5 (219064_at), score: 0.81 KCNK1potassium channel, subfamily K, member 1 (204679_at), score: 0.55 KCTD15potassium channel tetramerisation domain containing 15 (218553_s_at), score: 0.63 KIAA1644KIAA1644 (52837_at), score: 0.6 KLF5Kruppel-like factor 5 (intestinal) (209212_s_at), score: -0.74 LDB2LIM domain binding 2 (206481_s_at), score: -0.57 LEPRleptin receptor (209894_at), score: -0.66 LEPREL2leprecan-like 2 (204854_at), score: -0.65 LHPPphospholysine phosphohistidine inorganic pyrophosphate phosphatase (218523_at), score: -0.67 LMF1lipase maturation factor 1 (219135_s_at), score: -0.64 LMOD1leiomodin 1 (smooth muscle) (203766_s_at), score: -0.73 LOC100132540similar to LOC339047 protein (214870_x_at), score: 0.59 LOC339047hypothetical protein LOC339047 (221501_x_at), score: 0.61 LOC399491LOC399491 protein (214035_x_at), score: 0.58 LOC791120hypothetical LOC791120 (213367_at), score: 0.58 LPCAT4lysophosphatidylcholine acyltransferase 4 (40472_at), score: -0.66 LUZP1leucine zipper protein 1 (221832_s_at), score: -0.61 MATN2matrilin 2 (202350_s_at), score: -0.67 MR1major histocompatibility complex, class I-related (207565_s_at), score: -0.66 NBEAneurobeachin (221207_s_at), score: 0.54 NDUFA2NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 2, 8kDa (213550_s_at), score: -0.66 NME3non-metastatic cells 3, protein expressed in (204862_s_at), score: -0.77 NOTCH3Notch homolog 3 (Drosophila) (203238_s_at), score: -0.66 NPIPnuclear pore complex interacting protein (204538_x_at), score: 0.56 NRCAMneuronal cell adhesion molecule (204105_s_at), score: 0.89 OBFC1oligonucleotide/oligosaccharide-binding fold containing 1 (219100_at), score: -0.65 OCEL1occludin/ELL domain containing 1 (205441_at), score: -0.72 PALLDpalladin, cytoskeletal associated protein (200906_s_at), score: -0.66 PAPPA2pappalysin 2 (213332_at), score: 0.76 PCCApropionyl Coenzyme A carboxylase, alpha polypeptide (203860_at), score: -0.59 PCYOX1Lprenylcysteine oxidase 1 like (218953_s_at), score: -0.61 PDGFCplatelet derived growth factor C (218718_at), score: -0.63 PEX6peroxisomal biogenesis factor 6 (204545_at), score: -0.61 PFN2profilin 2 (204992_s_at), score: -0.58 PLGLB2plasminogen-like B2 (205871_at), score: 0.74 POLR2J4polymerase (RNA) II (DNA directed) polypeptide J4, pseudogene (60815_at), score: 0.56 PRELPproline/arginine-rich end leucine-rich repeat protein (204223_at), score: -0.76 PSMB9proteasome (prosome, macropain) subunit, beta type, 9 (large multifunctional peptidase 2) (204279_at), score: -0.66 PTERphosphotriesterase related (218967_s_at), score: 0.6 PYCARDPYD and CARD domain containing (221666_s_at), score: -0.57 RAGErenal tumor antigen (205130_at), score: -0.59 RALGPS2Ral GEF with PH domain and SH3 binding motif 2 (220338_at), score: 0.75 RGPD5RANBP2-like and GRIP domain containing 5 (210676_x_at), score: 0.57 SCN1Bsodium channel, voltage-gated, type I, beta (205508_at), score: -0.57 SLC17A5solute carrier family 17 (anion/sugar transporter), member 5 (221041_s_at), score: -0.59 SLC22A4solute carrier family 22 (organic cation/ergothioneine transporter), member 4 (205896_at), score: 0.56 SLC46A3solute carrier family 46, member 3 (214719_at), score: -0.58 SLC4A2solute carrier family 4, anion exchanger, member 2 (erythrocyte membrane protein band 3-like 1) (202111_at), score: 0.55 SLC6A9solute carrier family 6 (neurotransmitter transporter, glycine), member 9 (207043_s_at), score: 0.58 SPINT2serine peptidase inhibitor, Kunitz type, 2 (210715_s_at), score: -0.67 SPON1spondin 1, extracellular matrix protein (209436_at), score: 1 STAG1stromal antigen 1 (202293_at), score: 0.55 STARD5StAR-related lipid transfer (START) domain containing 5 (213820_s_at), score: -0.78 STAT4signal transducer and activator of transcription 4 (206118_at), score: -0.67 SUPT3Hsuppressor of Ty 3 homolog (S. cerevisiae) (206506_s_at), score: -0.62 SYNE1spectrin repeat containing, nuclear envelope 1 (209447_at), score: 0.56 TAP1transporter 1, ATP-binding cassette, sub-family B (MDR/TAP) (202307_s_at), score: -0.66 TAPBPTAP binding protein (tapasin) (208829_at), score: -0.6 TBL2transducin (beta)-like 2 (212685_s_at), score: 0.54 TBX1T-box 1 (211273_s_at), score: -0.7 TCF7L1transcription factor 7-like 1 (T-cell specific, HMG-box) (221016_s_at), score: -0.58 TCTN3tectonic family member 3 (212121_at), score: -0.57 THYN1thymocyte nuclear protein 1 (218491_s_at), score: -0.56 TJP2tight junction protein 2 (zona occludens 2) (202085_at), score: -0.56 TM4SF20transmembrane 4 L six family member 20 (220639_at), score: -0.58 TMEM176Btransmembrane protein 176B (220532_s_at), score: 0.57 TMEM187transmembrane protein 187 (204340_at), score: -0.69 TNFSF12tumor necrosis factor (ligand) superfamily, member 12 (205611_at), score: -0.56 TRAPPC6Atrafficking protein particle complex 6A (204985_s_at), score: -0.74 UBA7ubiquitin-like modifier activating enzyme 7 (203281_s_at), score: -0.69 UCP2uncoupling protein 2 (mitochondrial, proton carrier) (208998_at), score: -0.67 VAMP1vesicle-associated membrane protein 1 (synaptobrevin 1) (213326_at), score: -0.84 VCAM1vascular cell adhesion molecule 1 (203868_s_at), score: -0.64 VDRvitamin D (1,25- dihydroxyvitamin D3) receptor (204255_s_at), score: 0.63 ZMIZ1zinc finger, MIZ-type containing 1 (212124_at), score: 0.59 ZNF136zinc finger protein 136 (206240_s_at), score: 0.55 ZNF37Bzinc finger protein 37B (215358_x_at), score: 0.58
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| F348_WBS.CEL | 16 | 8 | WBS | hgu133plus2 | WBS | WBS 1 |
| F223_WBS.CEL | 15 | 8 | WBS | hgu133plus2 | WBS | WBS 1 |
| H652_WBS.CEL | 17 | 8 | WBS | hgu133plus2 | WBS | WBS 1 |
| F055_WBS.CEL | 14 | 8 | WBS | hgu133plus2 | WBS | WBS 1 |
| 10590_WBS.CEL | 2 | 8 | WBS | hgu133plus2 | WBS | WBS 1 |
| D890_WBS.CEL | 13 | 8 | WBS | hgu133plus2 | WBS | WBS 1 |
| 4319_WBS.CEL | 5 | 8 | WBS | hgu133plus2 | WBS | WBS 1 |
| E-GEOD-3860-raw-cel-1561690344.cel | 10 | 5 | HGPS | hgu133a | none | GM00038C |
| 5290_CNTL.CEL | 7 | 8 | WBS | hgu133plus2 | none | WBS 1 |