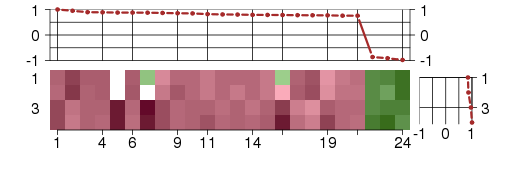

Under-expression is coded with green,
over-expression with red color.



ACER3alkaline ceramidase 3 (ENSG00000078124), score: 0.8 ARL14ADP-ribosylation factor-like 14 (ENSG00000179674), score: 0.95 AVPR1Aarginine vasopressin receptor 1A (ENSG00000166148), score: 0.82 CA6carbonic anhydrase VI (ENSG00000131686), score: 0.78 CTBSchitobiase, di-N-acetyl- (ENSG00000117151), score: 0.79 CYP39A1cytochrome P450, family 39, subfamily A, polypeptide 1 (ENSG00000146233), score: 0.88 DHTKD1dehydrogenase E1 and transketolase domain containing 1 (ENSG00000181192), score: 0.76 FETUBfetuin B (ENSG00000090512), score: 0.89 GAS6growth arrest-specific 6 (ENSG00000183087), score: -0.86 HALhistidine ammonia-lyase (ENSG00000084110), score: 0.85 IYDiodotyrosine deiodinase (ENSG00000009765), score: 0.79 KLHL5kelch-like 5 (Drosophila) (ENSG00000109790), score: -0.98 LECT2leukocyte cell-derived chemotaxin 2 (ENSG00000145826), score: 0.89 LRRC15leucine rich repeat containing 15 (ENSG00000172061), score: 0.88 MKLN1muskelin 1, intracellular mediator containing kelch motifs (ENSG00000128585), score: 0.77 NOP58NOP58 ribonucleoprotein homolog (yeast) (ENSG00000055044), score: 0.87 OTCornithine carbamoyltransferase (ENSG00000036473), score: 0.85 RGS18regulator of G-protein signaling 18 (ENSG00000150681), score: 0.77 SLC7A2solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 (ENSG00000003989), score: 0.78 STARD5StAR-related lipid transfer (START) domain containing 5 (ENSG00000172345), score: 0.81 TMEM135transmembrane protein 135 (ENSG00000166575), score: 0.76 TWSG1twisted gastrulation homolog 1 (Drosophila) (ENSG00000128791), score: -0.91 XKR9XK, Kell blood group complex subunit-related family, member 9 (ENSG00000221947), score: 1 ZC3H12Dzinc finger CCCH-type containing 12D (ENSG00000178199), score: 0.88
| Id | species | tissue | sex | individual |
|---|---|---|---|---|
| mdo_lv_m_ca1 | mdo | lv | m | _ |
| mdo_lv_f_ca1 | mdo | lv | f | _ |
| oan_lv_f_ca1 | oan | lv | f | _ |
| oan_lv_m_ca1 | oan | lv | m | _ |