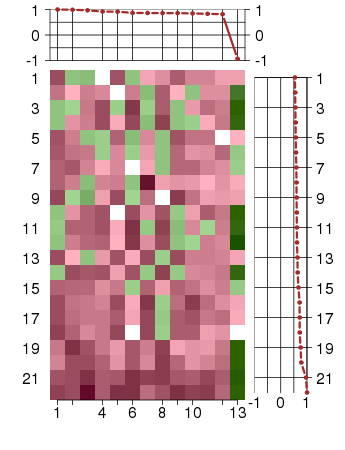

Under-expression is coded with green,
over-expression with red color.



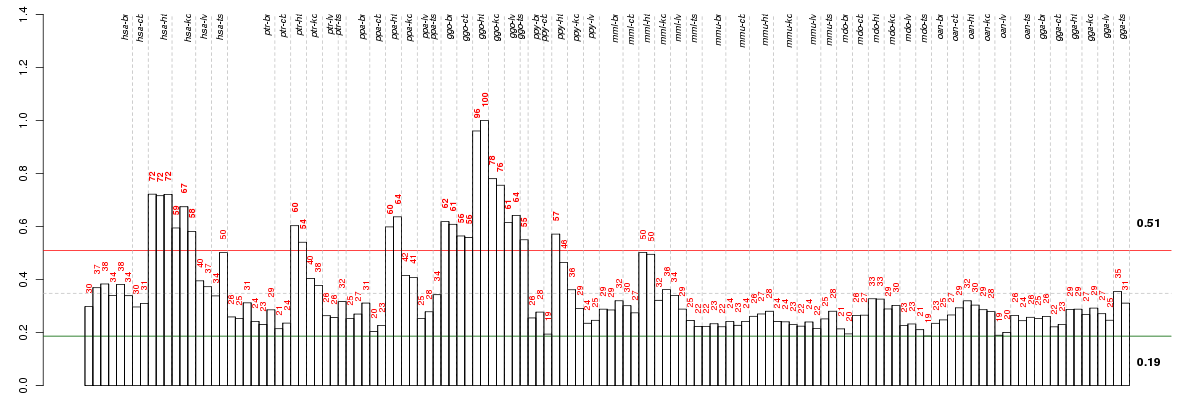
ANAPC16anaphase promoting complex subunit 16 (ENSG00000166295), score: 0.85 ANKRD2ankyrin repeat domain 2 (stretch responsive muscle) (ENSG00000165887), score: 0.86 C8orf45chromosome 8 open reading frame 45 (ENSG00000178460), score: 0.87 CACNA2D4calcium channel, voltage-dependent, alpha 2/delta subunit 4 (ENSG00000151062), score: 0.86 CCDC51coiled-coil domain containing 51 (ENSG00000164051), score: 0.81 CUL1cullin 1 (ENSG00000055130), score: -0.93 EMILIN3elastin microfibril interfacer 3 (ENSG00000183798), score: 0.99 ITGA11integrin, alpha 11 (ENSG00000137809), score: 0.92 MAPKAPK3mitogen-activated protein kinase-activated protein kinase 3 (ENSG00000114738), score: 0.91 MYOM3myomesin family, member 3 (ENSG00000142661), score: 1 PDAP1PDGFA associated protein 1 (ENSG00000106244), score: 0.83 PTCD1pentatricopeptide repeat domain 1 (ENSG00000106246), score: 0.97 SLC26A9solute carrier family 26, member 9 (ENSG00000174502), score: 0.86
| Id | species | tissue | sex | individual |
|---|---|---|---|---|
| ptr_ht_f_ca1 | ptr | ht | f | _ |
| ggo_ts_m_ca1 | ggo | ts | m | _ |
| ggo_cb_f_ca1 | ggo | cb | f | _ |
| ggo_cb_m_ca1 | ggo | cb | m | _ |
| ppy_ht_m_ca1 | ppy | ht | m | _ |
| hsa_kd_f_ca1 | hsa | kd | f | _ |
| hsa_kd_m2_ca1 | hsa | kd | m | 2 |
| ppa_ht_m_ca1 | ppa | ht | m | _ |
| ptr_ht_m_ca1 | ptr | ht | m | _ |
| ggo_br_f_ca1 | ggo | br | f | _ |
| ggo_lv_m_ca1 | ggo | lv | m | _ |
| ggo_br_m_ca1 | ggo | br | m | _ |
| ppa_ht_f_ca1 | ppa | ht | f | _ |
| ggo_lv_f_ca1 | ggo | lv | f | _ |
| hsa_kd_m1_ca1 | hsa | kd | m | 1 |
| hsa_ht_m2_ca1 | hsa | ht | m | 2 |
| hsa_ht_f_ca1 | hsa | ht | f | _ |
| hsa_ht_m1_ca1 | hsa | ht | m | 1 |
| ggo_kd_f_ca1 | ggo | kd | f | _ |
| ggo_kd_m_ca1 | ggo | kd | m | _ |
| ggo_ht_m_ca1 | ggo | ht | m | _ |
| ggo_ht_f_ca1 | ggo | ht | f | _ |