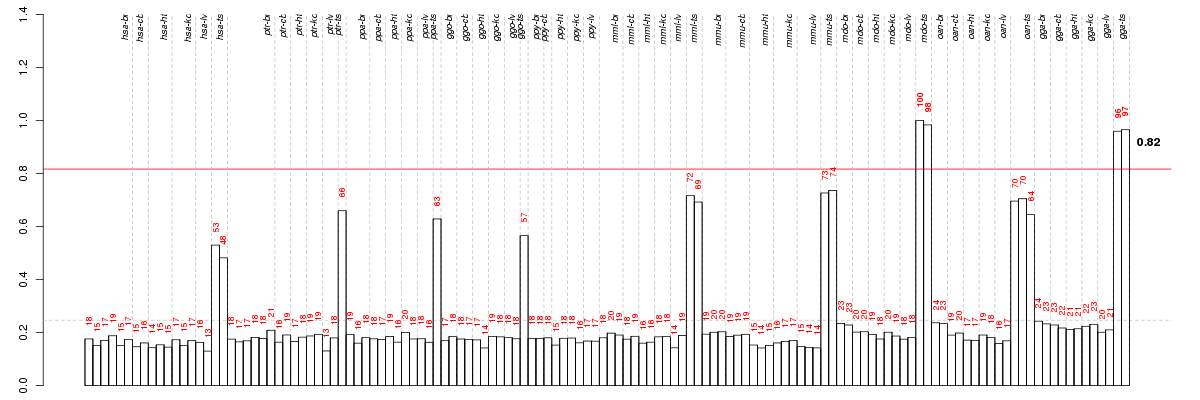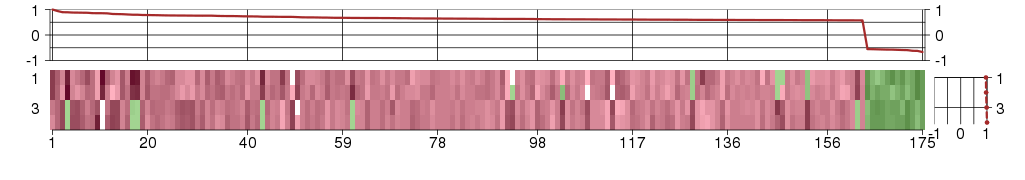

Under-expression is coded with green,
over-expression with red color.

chromosome segregation
The process by which genetic material, in the form of chromosomes, is organized into specific structures and then physically separated and apportioned to two or more sets.
M phase of mitotic cell cycle
A cell cycle process comprising the steps by which a cell progresses through M phase, the part of the mitotic cell cycle during which mitosis takes place.
mitotic cell cycle
Progression through the phases of the mitotic cell cycle, the most common eukaryotic cell cycle, which canonically comprises four successive phases called G1, S, G2, and M and includes replication of the genome and the subsequent segregation of chromosomes into daughter cells. In some variant cell cycles nuclear replication or nuclear division may not be followed by cell division, or G1 and G2 phases may be absent.
M phase
A cell cycle process comprising the steps by which a cell progresses through M phase, the part of the cell cycle comprising nuclear division.
nuclear division
A process by which a cell nucleus is divided into two nuclei, with DNA and other nuclear contents distributed between the daughter nuclei.
organelle organization
A process that is carried out at the cellular level which results in the assembly, arrangement of constituent parts, or disassembly of an organelle within a cell. An organelle is an organized structure of distinctive morphology and function. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton. Excludes the plasma membrane.
microtubule-based process
Any cellular process that depends upon or alters the microtubule cytoskeleton, that part of the cytoskeleton comprising microtubules and their associated proteins.
microtubule-based movement
Movement of organelles, other microtubules and other particles along microtubules, mediated by motor proteins.
cell cycle
The progression of biochemical and morphological phases and events that occur in a cell during successive cell replication or nuclear replication events. Canonically, the cell cycle comprises the replication and segregation of genetic material followed by the division of the cell, but in endocycles or syncytial cells nuclear replication or nuclear division may not be followed by cell division.
mitosis
A cell cycle process comprising the steps by which the nucleus of a eukaryotic cell divides; the process involves condensation of chromosomal DNA into a highly compacted form. Canonically, mitosis produces two daughter nuclei whose chromosome complement is identical to that of the mother cell.
biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
cellular process
Any process that is carried out at the cellular level, but not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
cellular component organization
A process that is carried out at the cellular level which results in the assembly, arrangement of constituent parts, or disassembly of a cellular component.
cell cycle process
A cellular process that is involved in the progression of biochemical and morphological phases and events that occur in a cell during successive cell replication or nuclear replication events.
cell cycle phase
A cell cycle process comprising the steps by which a cell progresses through one of the biochemical and morphological phases and events that occur during successive cell replication or nuclear replication events.
organelle fission
The creation of two or more organelles by division of one organelle.
all
NA
organelle organization
A process that is carried out at the cellular level which results in the assembly, arrangement of constituent parts, or disassembly of an organelle within a cell. An organelle is an organized structure of distinctive morphology and function. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton. Excludes the plasma membrane.
cell cycle process
A cellular process that is involved in the progression of biochemical and morphological phases and events that occur in a cell during successive cell replication or nuclear replication events.
mitosis
A cell cycle process comprising the steps by which the nucleus of a eukaryotic cell divides; the process involves condensation of chromosomal DNA into a highly compacted form. Canonically, mitosis produces two daughter nuclei whose chromosome complement is identical to that of the mother cell.
mitosis
A cell cycle process comprising the steps by which the nucleus of a eukaryotic cell divides; the process involves condensation of chromosomal DNA into a highly compacted form. Canonically, mitosis produces two daughter nuclei whose chromosome complement is identical to that of the mother cell.
M phase of mitotic cell cycle
A cell cycle process comprising the steps by which a cell progresses through M phase, the part of the mitotic cell cycle during which mitosis takes place.

condensed chromosome
A highly compacted molecule of DNA and associated proteins resulting in a cytologically distinct structure.
intracellular
The living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
chromosome, centromeric region
The region of a chromosome that includes the centromeric DNA and associated proteins. In monocentric chromosomes, this region corresponds to a single area of the chromosome, whereas in holocentric chromosomes, it is evenly distributed along the chromosome.
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
cell
The basic structural and functional unit of all organisms. Includes the plasma membrane and any external encapsulating structures such as the cell wall and cell envelope.
nucleus
A membrane-bounded organelle of eukaryotic cells in which chromosomes are housed and replicated. In most cells, the nucleus contains all of the cell's chromosomes except the organellar chromosomes, and is the site of RNA synthesis and processing. In some species, or in specialized cell types, RNA metabolism or DNA replication may be absent.
chromosome
A structure composed of a very long molecule of DNA and associated proteins (e.g. histones) that carries hereditary information.
spindle
The array of microtubules and associated molecules that forms between opposite poles of a eukaryotic cell during mitosis or meiosis and serves to move the duplicated chromosomes apart.
cytoskeleton
Any of the various filamentous elements that form the internal framework of cells, and typically remain after treatment of the cells with mild detergent to remove membrane constituents and soluble components of the cytoplasm. The term embraces intermediate filaments, microfilaments, microtubules, the microtrabecular lattice, and other structures characterized by a polymeric filamentous nature and long-range order within the cell. The various elements of the cytoskeleton not only serve in the maintenance of cellular shape but also have roles in other cellular functions, including cellular movement, cell division, endocytosis, and movement of organelles.
microtubule
Any of the long, generally straight, hollow tubes of internal diameter 12-15 nm and external diameter 24 nm found in a wide variety of eukaryotic cells; each consists (usually) of 13 protofilaments of polymeric tubulin, staggered in such a manner that the tubulin monomers are arranged in a helical pattern on the microtubular surface, and with the alpha/beta axes of the tubulin subunits parallel to the long axis of the tubule; exist in equilibrium with pool of tubulin monomers and can be rapidly assembled or disassembled in response to physiological stimuli; concerned with force generation, e.g. in the spindle.
microtubule cytoskeleton
The part of the cytoskeleton (the internal framework of a cell) composed of microtubules and associated proteins.
macromolecular complex
A stable assembly of two or more macromolecules, i.e. proteins, nucleic acids, carbohydrates or lipids, in which the constituent parts function together.
organelle
Organized structure of distinctive morphology and function. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton, and prokaryotic structures such as anammoxosomes and pirellulosomes. Excludes the plasma membrane.
membrane-bounded organelle
Organized structure of distinctive morphology and function, bounded by a single or double lipid bilayer membrane. Includes the nucleus, mitochondria, plastids, vacuoles, and vesicles. Excludes the plasma membrane.
non-membrane-bounded organelle
Organized structure of distinctive morphology and function, not bounded by a lipid bilayer membrane. Includes ribosomes, the cytoskeleton and chromosomes.
intracellular organelle
Organized structure of distinctive morphology and function, occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton. Excludes the plasma membrane.
intracellular membrane-bounded organelle
Organized structure of distinctive morphology and function, bounded by a single or double lipid bilayer membrane and occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, and vesicles. Excludes the plasma membrane.
intracellular non-membrane-bounded organelle
Organized structure of distinctive morphology and function, not bounded by a lipid bilayer membrane and occurring within the cell. Includes ribosomes, the cytoskeleton and chromosomes.
protein complex
Any macromolecular complex composed of two or more polypeptide subunits, which may or may not be identical. Protein complexes may have other associated non-protein prosthetic groups, such as nucleotides, metal ions or other small molecules.
organelle part
Any constituent part of an organelle, an organized structure of distinctive morphology and function. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton, but excludes the plasma membrane.
intracellular part
Any constituent part of the living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
chromosomal part
Any constituent part of a chromosome, a structure composed of a very long molecule of DNA and associated proteins (e.g. histones) that carries hereditary information.
cytoskeletal part
Any constituent part of the cytoskeleton, a cellular scaffolding or skeleton that maintains cell shape, enables some cell motion (using structures such as flagella and cilia), and plays important roles in both intra-cellular transport (e.g. the movement of vesicles and organelles) and cellular division. Includes constituent parts of intermediate filaments, microfilaments, microtubules, and the microtrabecular lattice.
intracellular organelle part
A constituent part of an intracellular organelle, an organized structure of distinctive morphology and function, occurring within the cell. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton but excludes the plasma membrane.
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
all
NA
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
organelle part
Any constituent part of an organelle, an organized structure of distinctive morphology and function. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton, but excludes the plasma membrane.
intracellular membrane-bounded organelle
Organized structure of distinctive morphology and function, bounded by a single or double lipid bilayer membrane and occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, and vesicles. Excludes the plasma membrane.
intracellular non-membrane-bounded organelle
Organized structure of distinctive morphology and function, not bounded by a lipid bilayer membrane and occurring within the cell. Includes ribosomes, the cytoskeleton and chromosomes.
intracellular organelle part
A constituent part of an intracellular organelle, an organized structure of distinctive morphology and function, occurring within the cell. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton but excludes the plasma membrane.
intracellular part
Any constituent part of the living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
intracellular organelle
Organized structure of distinctive morphology and function, occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton. Excludes the plasma membrane.
intracellular organelle part
A constituent part of an intracellular organelle, an organized structure of distinctive morphology and function, occurring within the cell. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton but excludes the plasma membrane.
spindle
The array of microtubules and associated molecules that forms between opposite poles of a eukaryotic cell during mitosis or meiosis and serves to move the duplicated chromosomes apart.
microtubule
Any of the long, generally straight, hollow tubes of internal diameter 12-15 nm and external diameter 24 nm found in a wide variety of eukaryotic cells; each consists (usually) of 13 protofilaments of polymeric tubulin, staggered in such a manner that the tubulin monomers are arranged in a helical pattern on the microtubular surface, and with the alpha/beta axes of the tubulin subunits parallel to the long axis of the tubule; exist in equilibrium with pool of tubulin monomers and can be rapidly assembled or disassembled in response to physiological stimuli; concerned with force generation, e.g. in the spindle.
chromosomal part
Any constituent part of a chromosome, a structure composed of a very long molecule of DNA and associated proteins (e.g. histones) that carries hereditary information.
cytoskeletal part
Any constituent part of the cytoskeleton, a cellular scaffolding or skeleton that maintains cell shape, enables some cell motion (using structures such as flagella and cilia), and plays important roles in both intra-cellular transport (e.g. the movement of vesicles and organelles) and cellular division. Includes constituent parts of intermediate filaments, microfilaments, microtubules, and the microtrabecular lattice.
spindle
The array of microtubules and associated molecules that forms between opposite poles of a eukaryotic cell during mitosis or meiosis and serves to move the duplicated chromosomes apart.
microtubule
Any of the long, generally straight, hollow tubes of internal diameter 12-15 nm and external diameter 24 nm found in a wide variety of eukaryotic cells; each consists (usually) of 13 protofilaments of polymeric tubulin, staggered in such a manner that the tubulin monomers are arranged in a helical pattern on the microtubular surface, and with the alpha/beta axes of the tubulin subunits parallel to the long axis of the tubule; exist in equilibrium with pool of tubulin monomers and can be rapidly assembled or disassembled in response to physiological stimuli; concerned with force generation, e.g. in the spindle.

molecular_function
Elemental activities, such as catalysis or binding, describing the actions of a gene product at the molecular level. A given gene product may exhibit one or more molecular functions.
nucleic acid binding
Interacting selectively and non-covalently with any nucleic acid.
DNA binding
Any molecular function by which a gene product interacts selectively with DNA (deoxyribonucleic acid).
chromatin binding
Interacting selectively and non-covalently with chromatin, the network of fibers of DNA and protein that make up the chromosomes of the eukaryotic nucleus during interphase.
motor activity
Catalysis of movement along a polymeric molecule such as a microfilament or microtubule, coupled to the hydrolysis of a nucleoside triphosphate.
microtubule motor activity
Catalysis of movement along a microtubule, coupled to the hydrolysis of a nucleoside triphosphate (usually ATP).
catalytic activity
Catalysis of a biochemical reaction at physiological temperatures. In biologically catalyzed reactions, the reactants are known as substrates, and the catalysts are naturally occurring macromolecular substances known as enzymes. Enzymes possess specific binding sites for substrates, and are usually composed wholly or largely of protein, but RNA that has catalytic activity (ribozyme) is often also regarded as enzymatic.
nucleoside-triphosphatase activity
Catalysis of the reaction: a nucleoside triphosphate + H2O = nucleoside diphosphate + phosphate.
binding
The selective, non-covalent, often stoichiometric, interaction of a molecule with one or more specific sites on another molecule.
pyrophosphatase activity
Catalysis of the hydrolysis of a pyrophosphate bond between two phosphate groups, leaving one phosphate on each of the two fragments.
hydrolase activity
Catalysis of the hydrolysis of various bonds, e.g. C-O, C-N, C-C, phosphoric anhydride bonds, etc. Hydrolase is the systematic name for any enzyme of EC class 3.
hydrolase activity, acting on acid anhydrides
Catalysis of the hydrolysis of any acid anhydride.
hydrolase activity, acting on acid anhydrides, in phosphorus-containing anhydrides
Catalysis of the hydrolysis of any acid anhydride which contains phosphorus.
all
NA
ACER1alkaline ceramidase 1 (ENSG00000167769), score: 0.88 ADSLadenylosuccinate lyase (ENSG00000100354), score: 0.61 AGBL3ATP/GTP binding protein-like 3 (ENSG00000146856), score: 0.67 AIREautoimmune regulator (ENSG00000160224), score: 0.82 ANKFN1ankyrin-repeat and fibronectin type III domain containing 1 (ENSG00000153930), score: 0.7 ANXA6annexin A6 (ENSG00000197043), score: -0.57 ANXA7annexin A7 (ENSG00000138279), score: -0.55 APTXaprataxin (ENSG00000137074), score: 0.88 ASB7ankyrin repeat and SOCS box-containing 7 (ENSG00000183475), score: 0.68 ASPMasp (abnormal spindle) homolog, microcephaly associated (Drosophila) (ENSG00000066279), score: 0.62 B3GNT5UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 5 (ENSG00000176597), score: 0.61 BBS7Bardet-Biedl syndrome 7 (ENSG00000138686), score: 0.6 BRPF1bromodomain and PHD finger containing, 1 (ENSG00000156983), score: 0.66 C12orf50chromosome 12 open reading frame 50 (ENSG00000165805), score: 0.65 C12orf63chromosome 12 open reading frame 63 (ENSG00000188596), score: 0.77 C13orf34chromosome 13 open reading frame 34 (ENSG00000136122), score: 0.57 C14orf166Bchromosome 14 open reading frame 166B (ENSG00000100565), score: 0.64 C14orf39chromosome 14 open reading frame 39 (ENSG00000179008), score: 0.62 C15orf26chromosome 15 open reading frame 26 (ENSG00000156206), score: 0.64 C17orf104chromosome 17 open reading frame 104 (ENSG00000180336), score: 0.73 C19orf45chromosome 19 open reading frame 45 (ENSG00000198723), score: 0.59 C1orf100chromosome 1 open reading frame 100 (ENSG00000173728), score: 0.59 C1orf130chromosome 1 open reading frame 130 (ENSG00000184454), score: 0.6 C1orf158chromosome 1 open reading frame 158 (ENSG00000157330), score: 0.58 C1orf9chromosome 1 open reading frame 9 (ENSG00000094975), score: 0.72 C20orf79chromosome 20 open reading frame 79 (ENSG00000132631), score: 0.58 C20orf85chromosome 20 open reading frame 85 (ENSG00000124237), score: 0.63 C3orf19chromosome 3 open reading frame 19 (ENSG00000154781), score: 0.6 C4orf47chromosome 4 open reading frame 47 (ENSG00000205129), score: 0.61 C6orf150chromosome 6 open reading frame 150 (ENSG00000164430), score: 0.59 C6orf204chromosome 6 open reading frame 204 (ENSG00000111860), score: 0.58 C7orf45chromosome 7 open reading frame 45 (ENSG00000165120), score: 0.71 C9orf82chromosome 9 open reading frame 82 (ENSG00000120159), score: 0.67 C9orf98chromosome 9 open reading frame 98 (ENSG00000165695), score: 0.6 CCDC108coiled-coil domain containing 108 (ENSG00000181378), score: 0.65 CCDC135coiled-coil domain containing 135 (ENSG00000159625), score: 0.63 CCDC27coiled-coil domain containing 27 (ENSG00000162592), score: 0.71 CCDC37coiled-coil domain containing 37 (ENSG00000163885), score: 0.73 CCDC40coiled-coil domain containing 40 (ENSG00000141519), score: 0.61 CCDC41coiled-coil domain containing 41 (ENSG00000173588), score: 0.59 CCDC43coiled-coil domain containing 43 (ENSG00000180329), score: 0.58 CCDC63coiled-coil domain containing 63 (ENSG00000173093), score: 0.63 CCDC67coiled-coil domain containing 67 (ENSG00000165325), score: 0.74 CDC42BPBCDC42 binding protein kinase beta (DMPK-like) (ENSG00000198752), score: -0.56 CENPIcentromere protein I (ENSG00000102384), score: 0.75 CENPKcentromere protein K (ENSG00000123219), score: 0.63 CENPQcentromere protein Q (ENSG00000031691), score: 0.65 CEP152centrosomal protein 152kDa (ENSG00000103995), score: 0.65 CEP55centrosomal protein 55kDa (ENSG00000138180), score: 0.62 CNOT4CCR4-NOT transcription complex, subunit 4 (ENSG00000080802), score: 0.61 CNOT8CCR4-NOT transcription complex, subunit 8 (ENSG00000155508), score: 0.76 CRYBA1crystallin, beta A1 (ENSG00000108255), score: 0.8 CTTNcortactin (ENSG00000085733), score: -0.57 CXorf22chromosome X open reading frame 22 (ENSG00000165164), score: 1 CXorf30chromosome X open reading frame 30 (ENSG00000205081), score: 0.82 DIS3DIS3 mitotic control homolog (S. cerevisiae) (ENSG00000083520), score: 0.58 DMC1DMC1 dosage suppressor of mck1 homolog, meiosis-specific homologous recombination (yeast) (ENSG00000100206), score: 0.59 E2F8E2F transcription factor 8 (ENSG00000129173), score: 0.94 ECDecdysoneless homolog (Drosophila) (ENSG00000122882), score: 0.58 EFCAB1EF-hand calcium binding domain 1 (ENSG00000034239), score: 0.59 EFCAB5EF-hand calcium binding domain 5 (ENSG00000176927), score: 0.66 EFNB2ephrin-B2 (ENSG00000125266), score: -0.57 ENO4enolase family member 4 (ENSG00000188316), score: 0.78 ENPP4ectonucleotide pyrophosphatase/phosphodiesterase 4 (putative) (ENSG00000001561), score: 0.77 FAM54Afamily with sequence similarity 54, member A (ENSG00000146410), score: 0.73 FAM83Dfamily with sequence similarity 83, member D (ENSG00000101447), score: 0.63 FBXO43F-box protein 43 (ENSG00000156509), score: 0.62 FKBP14FK506 binding protein 14, 22 kDa (ENSG00000106080), score: 0.76 FNDC3Afibronectin type III domain containing 3A (ENSG00000102531), score: 0.67 FOXN1forkhead box N1 (ENSG00000109101), score: 0.86 FSD1Lfibronectin type III and SPRY domain containing 1-like (ENSG00000106701), score: 0.59 GAS8growth arrest-specific 8 (ENSG00000141013), score: 0.6 GDPD4glycerophosphodiester phosphodiesterase domain containing 4 (ENSG00000178795), score: 0.63 GEMC1geminin coiled-coil domain-containing protein 1 (ENSG00000205835), score: 0.77 GEMIN5gem (nuclear organelle) associated protein 5 (ENSG00000082516), score: 0.58 GEMIN6gem (nuclear organelle) associated protein 6 (ENSG00000152147), score: 0.58 GLI3GLI family zinc finger 3 (ENSG00000106571), score: 0.66 GM2AGM2 ganglioside activator (ENSG00000196743), score: -0.56 GORASP1golgi reassembly stacking protein 1, 65kDa (ENSG00000114745), score: -0.59 GPR1G protein-coupled receptor 1 (ENSG00000183671), score: 0.76 HEATR6HEAT repeat containing 6 (ENSG00000068097), score: 0.76 HELLShelicase, lymphoid-specific (ENSG00000119969), score: 0.64 HORMAD1HORMA domain containing 1 (ENSG00000143452), score: 0.59 HORMAD2HORMA domain containing 2 (ENSG00000176635), score: 0.63 HOXB13homeobox B13 (ENSG00000159184), score: 0.58 IFT74intraflagellar transport 74 homolog (Chlamydomonas) (ENSG00000096872), score: 0.65 IQUBIQ motif and ubiquitin domain containing (ENSG00000164675), score: 0.59 IWS1IWS1 homolog (S. cerevisiae) (ENSG00000163166), score: 0.59 KATNAL2katanin p60 subunit A-like 2 (ENSG00000167216), score: 0.6 KDM1Blysine (K)-specific demethylase 1B (ENSG00000165097), score: -0.62 KIAA1609KIAA1609 (ENSG00000140950), score: 0.71 KIF14kinesin family member 14 (ENSG00000118193), score: 0.67 KIF18Akinesin family member 18A (ENSG00000121621), score: 0.61 KIF18Bkinesin family member 18B (ENSG00000186185), score: 0.87 KIF24kinesin family member 24 (ENSG00000186638), score: 0.86 KIF27kinesin family member 27 (ENSG00000165115), score: 0.66 KIF2Bkinesin family member 2B (ENSG00000141200), score: 0.67 KLHL10kelch-like 10 (Drosophila) (ENSG00000161594), score: 0.69 KNTC1kinetochore associated 1 (ENSG00000184445), score: 0.58 KPNA6karyopherin alpha 6 (importin alpha 7) (ENSG00000025800), score: 0.61 LCA5Leber congenital amaurosis 5 (ENSG00000135338), score: 0.69 LCTlactase (ENSG00000115850), score: 0.62 LHCGRluteinizing hormone/choriogonadotropin receptor (ENSG00000138039), score: 0.61 LRRC48leucine rich repeat containing 48 (ENSG00000171962), score: 0.61 LRRC67leucine rich repeat containing 67 (ENSG00000178125), score: 0.63 LRRIQ4leucine-rich repeats and IQ motif containing 4 (ENSG00000188306), score: 0.66 MAKmale germ cell-associated kinase (ENSG00000111837), score: 0.64 MAP9microtubule-associated protein 9 (ENSG00000164114), score: 0.59 MCM9minichromosome maintenance complex component 9 (ENSG00000111877), score: 0.58 MELKmaternal embryonic leucine zipper kinase (ENSG00000165304), score: 0.6 MYBv-myb myeloblastosis viral oncogene homolog (avian) (ENSG00000118513), score: 0.89 MYBL1v-myb myeloblastosis viral oncogene homolog (avian)-like 1 (ENSG00000185697), score: 0.72 MYF5myogenic factor 5 (ENSG00000111049), score: 0.89 MYF6myogenic factor 6 (herculin) (ENSG00000111046), score: 0.72 NCAPGnon-SMC condensin I complex, subunit G (ENSG00000109805), score: 0.6 NPSR1neuropeptide S receptor 1 (ENSG00000187258), score: 0.61 NR5A1nuclear receptor subfamily 5, group A, member 1 (ENSG00000136931), score: 0.58 NUF2NUF2, NDC80 kinetochore complex component, homolog (S. cerevisiae) (ENSG00000143228), score: 0.62 NUP54nucleoporin 54kDa (ENSG00000138750), score: 0.78 ORC1Lorigin recognition complex, subunit 1-like (S. cerevisiae) (ENSG00000085840), score: 0.78 PAPD4PAP associated domain containing 4 (ENSG00000164329), score: 0.69 PARK7Parkinson disease (autosomal recessive, early onset) 7 (ENSG00000116288), score: -0.59 PDIK1LPDLIM1 interacting kinase 1 like (ENSG00000175087), score: 0.6 PHTF1putative homeodomain transcription factor 1 (ENSG00000116793), score: 0.67 PKD2L2polycystic kidney disease 2-like 2 (ENSG00000078795), score: 0.57 PLCL2phospholipase C-like 2 (ENSG00000154822), score: -0.58 PPEF1protein phosphatase, EF-hand calcium binding domain 1 (ENSG00000086717), score: 0.61 PPP1R8protein phosphatase 1, regulatory (inhibitor) subunit 8 (ENSG00000117751), score: 0.61 PRSSL1protease, serine-like 1 (ENSG00000185198), score: 0.6 PUS3pseudouridylate synthase 3 (ENSG00000110060), score: 0.87 RAB38RAB38, member RAS oncogene family (ENSG00000123892), score: 0.64 RAG2recombination activating gene 2 (ENSG00000175097), score: 0.67 RBM18RNA binding motif protein 18 (ENSG00000119446), score: 0.58 RBM46RNA binding motif protein 46 (ENSG00000151962), score: 0.66 RFX3regulatory factor X, 3 (influences HLA class II expression) (ENSG00000080298), score: 0.68 RFX6regulatory factor X, 6 (ENSG00000185002), score: 0.85 RGP1RGP1 retrograde golgi transport homolog (S. cerevisiae) (ENSG00000107185), score: 0.62 RIBC2RIB43A domain with coiled-coils 2 (ENSG00000128408), score: 0.58 RNF17ring finger protein 17 (ENSG00000132972), score: 0.63 RPE65retinal pigment epithelium-specific protein 65kDa (ENSG00000116745), score: 0.76 RSPH4Aradial spoke head 4 homolog A (Chlamydomonas) (ENSG00000111834), score: 0.57 SGOL1shugoshin-like 1 (S. pombe) (ENSG00000129810), score: 0.78 SKILSKI-like oncogene (ENSG00000136603), score: 0.59 SMARCAD1SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1 (ENSG00000163104), score: 0.6 SMC1Bstructural maintenance of chromosomes 1B (ENSG00000077935), score: 0.74 SPATA18spermatogenesis associated 18 homolog (rat) (ENSG00000163071), score: 0.59 STILSCL/TAL1 interrupting locus (ENSG00000123473), score: 0.76 STRA8stimulated by retinoic acid gene 8 homolog (mouse) (ENSG00000146857), score: 0.76 TDRD5tudor domain containing 5 (ENSG00000162782), score: 0.69 TEKT3tektin 3 (ENSG00000125409), score: 0.67 TERTtelomerase reverse transcriptase (ENSG00000164362), score: 0.82 THAP1THAP domain containing, apoptosis associated protein 1 (ENSG00000131931), score: 0.64 TLX1T-cell leukemia homeobox 1 (ENSG00000107807), score: 0.8 TMEM26transmembrane protein 26 (ENSG00000196932), score: 0.74 TMPRSS7transmembrane protease, serine 7 (ENSG00000176040), score: 0.63 TNPO3transportin 3 (ENSG00000064419), score: 0.59 TOP2Atopoisomerase (DNA) II alpha 170kDa (ENSG00000131747), score: 0.67 TP53I11tumor protein p53 inducible protein 11 (ENSG00000175274), score: -0.67 TP53TG5TP53 target 5 (ENSG00000124251), score: 0.76 TP63tumor protein p63 (ENSG00000073282), score: 0.71 TPX2TPX2, microtubule-associated, homolog (Xenopus laevis) (ENSG00000088325), score: 0.59 TRAF3IP1TNF receptor-associated factor 3 interacting protein 1 (ENSG00000204104), score: 0.64 TRIM71tripartite motif-containing 71 (ENSG00000206557), score: 0.63 TSGA14testis specific, 14 (ENSG00000106477), score: 0.68 UHRF1ubiquitin-like with PHD and ring finger domains 1 (ENSG00000034063), score: 0.81 VASH2vasohibin 2 (ENSG00000143494), score: 0.73 VWA5B1von Willebrand factor A domain containing 5B1 (ENSG00000158816), score: 0.62 WDR64WD repeat domain 64 (ENSG00000162843), score: 0.65 XPO4exportin 4 (ENSG00000132953), score: 0.59 YME1L1YME1-like 1 (S. cerevisiae) (ENSG00000136758), score: 0.58 ZMYND11zinc finger, MYND domain containing 11 (ENSG00000015171), score: -0.62 ZNF512zinc finger protein 512 (ENSG00000243943), score: 0.6 ZNRF4zinc and ring finger 4 (ENSG00000105428), score: 0.64
| Id | species | tissue | sex | individual |
|---|---|---|---|---|
| gga_ts_m1_ca1 | gga | ts | m | 1 |
| gga_ts_m2_ca1 | gga | ts | m | 2 |
| mdo_ts_m1_ca1 | mdo | ts | m | 1 |
| mdo_ts_m2_ca1 | mdo | ts | m | 2 |