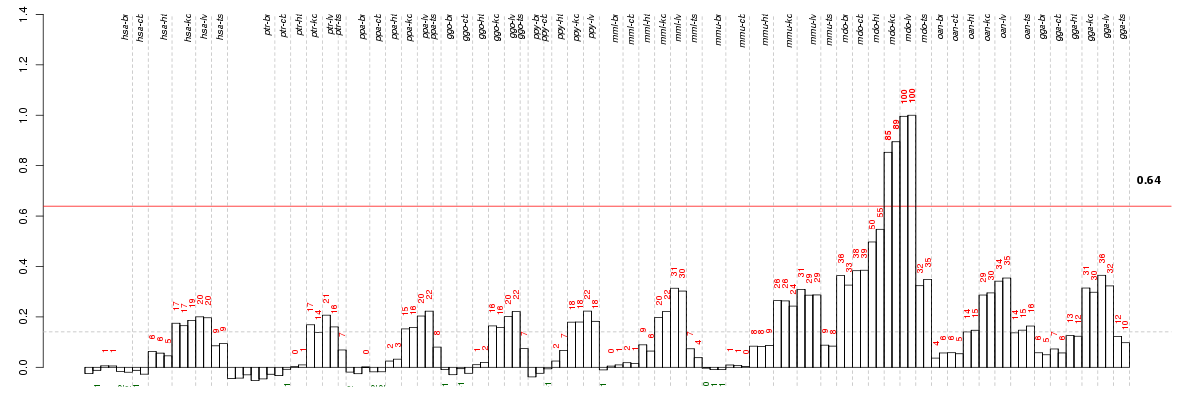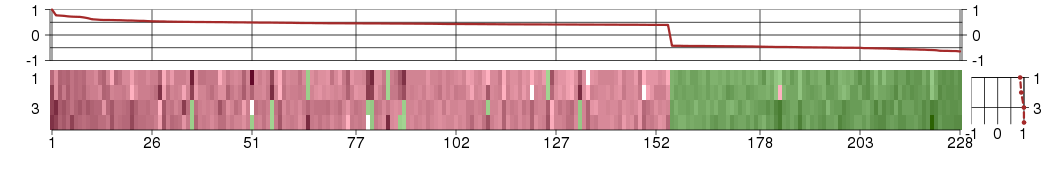

Under-expression is coded with green,
over-expression with red color.



AARS2alanyl-tRNA synthetase 2, mitochondrial (putative) (ENSG00000124608), score: 0.5 ABCA5ATP-binding cassette, sub-family A (ABC1), member 5 (ENSG00000154265), score: -0.47 ABCD3ATP-binding cassette, sub-family D (ALD), member 3 (ENSG00000117528), score: 0.42 ABCG1ATP-binding cassette, sub-family G (WHITE), member 1 (ENSG00000160179), score: -0.5 ABHD6abhydrolase domain containing 6 (ENSG00000163686), score: -0.57 ACBD5acyl-CoA binding domain containing 5 (ENSG00000107897), score: 0.41 ACER2alkaline ceramidase 2 (ENSG00000177076), score: 0.52 ACER3alkaline ceramidase 3 (ENSG00000078124), score: 0.49 ACO1aconitase 1, soluble (ENSG00000122729), score: 0.42 ACP2acid phosphatase 2, lysosomal (ENSG00000134575), score: 0.4 ACTL6Aactin-like 6A (ENSG00000136518), score: 0.4 ACYP2acylphosphatase 2, muscle type (ENSG00000170634), score: -0.62 ADCK1aarF domain containing kinase 1 (ENSG00000063761), score: -0.54 AGXTalanine-glyoxylate aminotransferase (ENSG00000172482), score: 0.44 AHRaryl hydrocarbon receptor (ENSG00000106546), score: 0.4 AIFM2apoptosis-inducing factor, mitochondrion-associated, 2 (ENSG00000042286), score: 0.46 AK2adenylate kinase 2 (ENSG00000004455), score: 0.42 AMBRA1autophagy/beclin-1 regulator 1 (ENSG00000110497), score: 0.52 ANAPC4anaphase promoting complex subunit 4 (ENSG00000053900), score: -0.56 ANKRD22ankyrin repeat domain 22 (ENSG00000152766), score: 0.46 AP1G1adaptor-related protein complex 1, gamma 1 subunit (ENSG00000166747), score: 0.4 AP2B1adaptor-related protein complex 2, beta 1 subunit (ENSG00000006125), score: -0.49 APEHN-acylaminoacyl-peptide hydrolase (ENSG00000164062), score: 0.44 ARFGAP3ADP-ribosylation factor GTPase activating protein 3 (ENSG00000242247), score: 0.5 ARHGEF38Rho guanine nucleotide exchange factor (GEF) 38 (ENSG00000138784), score: 1 ARL14ADP-ribosylation factor-like 14 (ENSG00000179674), score: 0.57 ARSKarylsulfatase family, member K (ENSG00000164291), score: 0.47 ASXL2additional sex combs like 2 (Drosophila) (ENSG00000143970), score: 0.43 ATG4AATG4 autophagy related 4 homolog A (S. cerevisiae) (ENSG00000101844), score: 0.43 ATMINATM interactor (ENSG00000166454), score: -0.58 ATP13A3ATPase type 13A3 (ENSG00000133657), score: 0.51 ATP2B4ATPase, Ca++ transporting, plasma membrane 4 (ENSG00000058668), score: -0.55 ATP2C1ATPase, Ca++ transporting, type 2C, member 1 (ENSG00000017260), score: -0.48 ATXN2ataxin 2 (ENSG00000204842), score: 0.5 AVPR1Aarginine vasopressin receptor 1A (ENSG00000166148), score: 0.46 C12orf66chromosome 12 open reading frame 66 (ENSG00000174206), score: 0.48 C17orf75chromosome 17 open reading frame 75 (ENSG00000108666), score: 0.43 C1orf116chromosome 1 open reading frame 116 (ENSG00000182795), score: 0.45 C1orf123chromosome 1 open reading frame 123 (ENSG00000162384), score: 0.4 C20orf43chromosome 20 open reading frame 43 (ENSG00000022277), score: -0.48 CAMSAP1calmodulin regulated spectrin-associated protein 1 (ENSG00000130559), score: -0.43 CAPRIN2caprin family member 2 (ENSG00000110888), score: -0.53 CASS4Cas scaffolding protein family member 4 (ENSG00000087589), score: 0.46 CCDC82coiled-coil domain containing 82 (ENSG00000149231), score: -0.57 CDC123cell division cycle 123 homolog (S. cerevisiae) (ENSG00000151465), score: -0.52 CDCA3cell division cycle associated 3 (ENSG00000111665), score: 0.51 CEPT1choline/ethanolamine phosphotransferase 1 (ENSG00000134255), score: 0.42 CLDN19claudin 19 (ENSG00000164007), score: 0.45 CLTBclathrin, light chain B (ENSG00000175416), score: -0.63 CNGA2cyclic nucleotide gated channel alpha 2 (ENSG00000183862), score: 0.41 COX1cytochrome c oxidase subunit I (ENSG00000198804), score: -0.56 CREB3L3cAMP responsive element binding protein 3-like 3 (ENSG00000060566), score: 0.52 CRYGNcrystallin, gamma N (ENSG00000127377), score: 0.52 CTBSchitobiase, di-N-acetyl- (ENSG00000117151), score: 0.55 CTHcystathionase (cystathionine gamma-lyase) (ENSG00000116761), score: 0.43 CYLDcylindromatosis (turban tumor syndrome) (ENSG00000083799), score: -0.42 CYP39A1cytochrome P450, family 39, subfamily A, polypeptide 1 (ENSG00000146233), score: 0.42 CYTBcytochrome b (ENSG00000198727), score: -0.65 CYTH1cytohesin 1 (ENSG00000108669), score: -0.56 DCTN5dynactin 5 (p25) (ENSG00000166847), score: -0.5 DCUN1D4DCN1, defective in cullin neddylation 1, domain containing 4 (S. cerevisiae) (ENSG00000109184), score: -0.6 DHDPSLdihydrodipicolinate synthase-like, mitochondrial (ENSG00000241935), score: 0.44 DHRS7Bdehydrogenase/reductase (SDR family) member 7B (ENSG00000109016), score: -0.5 EEA1early endosome antigen 1 (ENSG00000102189), score: 0.56 EIF2AK3eukaryotic translation initiation factor 2-alpha kinase 3 (ENSG00000172071), score: 0.4 EIF3Meukaryotic translation initiation factor 3, subunit M (ENSG00000149100), score: 0.51 ELF5E74-like factor 5 (ets domain transcription factor) (ENSG00000135374), score: 0.5 ELMO3engulfment and cell motility 3 (ENSG00000102890), score: 0.42 EPHA1EPH receptor A1 (ENSG00000146904), score: 0.46 EPYCepiphycan (ENSG00000083782), score: 0.53 ERCC6excision repair cross-complementing rodent repair deficiency, complementation group 6 (ENSG00000225830), score: -0.47 ERICH1glutamate-rich 1 (ENSG00000104714), score: -0.44 EXD2exonuclease 3'-5' domain containing 2 (ENSG00000081177), score: -0.45 EXOC3exocyst complex component 3 (ENSG00000180104), score: -0.43 EXOSC9exosome component 9 (ENSG00000123737), score: 0.41 FAM110Cfamily with sequence similarity 110, member C (ENSG00000184731), score: 0.42 FAM118Bfamily with sequence similarity 118, member B (ENSG00000197798), score: -0.45 FAM18Afamily with sequence similarity 18, member A (ENSG00000166676), score: -0.49 FAM19A4family with sequence similarity 19 (chemokine (C-C motif)-like), member A4 (ENSG00000163377), score: 0.42 FAM3Dfamily with sequence similarity 3, member D (ENSG00000198643), score: 0.75 FAM82A2family with sequence similarity 82, member A2 (ENSG00000137824), score: 0.66 FAM82Bfamily with sequence similarity 82, member B (ENSG00000176623), score: 0.4 FBXO3F-box protein 3 (ENSG00000110429), score: 0.41 FERMT1fermitin family member 1 (ENSG00000101311), score: 0.51 FRA10AC1fragile site, folic acid type, rare, fra(10)(q23.3) or fra(10)(q24.2) candidate 1 (ENSG00000148690), score: -0.44 FUT10fucosyltransferase 10 (alpha (1,3) fucosyltransferase) (ENSG00000172728), score: 0.48 GABRPgamma-aminobutyric acid (GABA) A receptor, pi (ENSG00000094755), score: 0.49 GCM1glial cells missing homolog 1 (Drosophila) (ENSG00000137270), score: 0.46 GCNT7glucosaminyl (N-acetyl) transferase family member 7 (ENSG00000124091), score: 0.46 GK5glycerol kinase 5 (putative) (ENSG00000175066), score: 0.61 GLIPR2GLI pathogenesis-related 2 (ENSG00000122694), score: -0.44 GOPCgolgi-associated PDZ and coiled-coil motif containing (ENSG00000047932), score: -0.43 GRAMD2GRAM domain containing 2 (ENSG00000175318), score: 0.45 GSCgoosecoid homeobox (ENSG00000133937), score: 0.49 GXYLT1glucoside xylosyltransferase 1 (ENSG00000151233), score: 0.4 ICOSLGinducible T-cell co-stimulator ligand (ENSG00000160223), score: 0.53 IFT46intraflagellar transport 46 homolog (Chlamydomonas) (ENSG00000118096), score: -0.47 IGF1insulin-like growth factor 1 (somatomedin C) (ENSG00000017427), score: 0.41 IKIK cytokine, down-regulator of HLA II (ENSG00000113141), score: -0.52 IL6STinterleukin 6 signal transducer (gp130, oncostatin M receptor) (ENSG00000134352), score: 0.4 IMPAD1inositol monophosphatase domain containing 1 (ENSG00000104331), score: -0.59 IPPintracisternal A particle-promoted polypeptide (ENSG00000197429), score: 0.46 IQGAP1IQ motif containing GTPase activating protein 1 (ENSG00000140575), score: 0.44 KBTBD4kelch repeat and BTB (POZ) domain containing 4 (ENSG00000123444), score: -0.43 KIAA1432KIAA1432 (ENSG00000107036), score: 0.44 KIAA1826KIAA1826 (ENSG00000170903), score: -0.44 KIFAP3kinesin-associated protein 3 (ENSG00000075945), score: -0.49 KLHL5kelch-like 5 (Drosophila) (ENSG00000109790), score: -0.47 LAG3lymphocyte-activation gene 3 (ENSG00000089692), score: 0.41 LEPRE1leucine proline-enriched proteoglycan (leprecan) 1 (ENSG00000117385), score: 0.43 LGR4leucine-rich repeat-containing G protein-coupled receptor 4 (ENSG00000205213), score: 0.41 LOC100292202similar to solute carrier family 25, member 25 (ENSG00000148339), score: 0.41 LRATlecithin retinol acyltransferase (phosphatidylcholine--retinol O-acyltransferase) (ENSG00000121207), score: 0.42 LY75lymphocyte antigen 75 (ENSG00000054219), score: 0.41 LYSMD3LysM, putative peptidoglycan-binding, domain containing 3 (ENSG00000176018), score: 0.46 MARVELD3MARVEL domain containing 3 (ENSG00000140832), score: 0.5 MGAT4Amannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme A (ENSG00000071073), score: -0.45 MPP1membrane protein, palmitoylated 1, 55kDa (ENSG00000130830), score: 0.53 MPZL2myelin protein zero-like 2 (ENSG00000149573), score: 0.43 MRPL19mitochondrial ribosomal protein L19 (ENSG00000115364), score: 0.58 MTMR14myotubularin related protein 14 (ENSG00000163719), score: 0.71 MYH10myosin, heavy chain 10, non-muscle (ENSG00000133026), score: -0.53 N4BP1NEDD4 binding protein 1 (ENSG00000102921), score: 0.49 NADSYN1NAD synthetase 1 (ENSG00000172890), score: 0.54 NAGAN-acetylgalactosaminidase, alpha- (ENSG00000198951), score: 0.49 NCSTNnicastrin (ENSG00000162736), score: 0.41 ND2MTND2 (ENSG00000198763), score: -0.62 NDST4N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 4 (ENSG00000138653), score: 0.5 NECAB3N-terminal EF-hand calcium binding protein 3 (ENSG00000125967), score: -0.48 NOD1nucleotide-binding oligomerization domain containing 1 (ENSG00000106100), score: 0.47 NOXO1NADPH oxidase organizer 1 (ENSG00000196408), score: 0.71 NRBF2nuclear receptor binding factor 2 (ENSG00000148572), score: 0.51 NSUN2NOP2/Sun domain family, member 2 (ENSG00000037474), score: -0.49 NUDT12nudix (nucleoside diphosphate linked moiety X)-type motif 12 (ENSG00000112874), score: 0.46 OLFM4olfactomedin 4 (ENSG00000102837), score: 0.43 OSBPL2oxysterol binding protein-like 2 (ENSG00000130703), score: -0.44 OSBPL3oxysterol binding protein-like 3 (ENSG00000070882), score: 0.59 P4HTMprolyl 4-hydroxylase, transmembrane (endoplasmic reticulum) (ENSG00000178467), score: -0.52 PAFAH2platelet-activating factor acetylhydrolase 2, 40kDa (ENSG00000158006), score: 0.42 PCK1phosphoenolpyruvate carboxykinase 1 (soluble) (ENSG00000124253), score: 0.43 PCK2phosphoenolpyruvate carboxykinase 2 (mitochondrial) (ENSG00000100889), score: 0.41 PDP2pyruvate dehyrogenase phosphatase catalytic subunit 2 (ENSG00000172840), score: 0.59 PEX7peroxisomal biogenesis factor 7 (ENSG00000112357), score: 0.47 PFKFB36-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 (ENSG00000170525), score: -0.46 PHF12PHD finger protein 12 (ENSG00000109118), score: 0.41 PIGKphosphatidylinositol glycan anchor biosynthesis, class K (ENSG00000142892), score: 0.4 PIGLphosphatidylinositol glycan anchor biosynthesis, class L (ENSG00000108474), score: 0.54 PM20D1peptidase M20 domain containing 1 (ENSG00000162877), score: 0.45 PODNpodocan (ENSG00000174348), score: 0.62 POLR2Cpolymerase (RNA) II (DNA directed) polypeptide C, 33kDa (ENSG00000102978), score: -0.46 PPEF2protein phosphatase, EF-hand calcium binding domain 2 (ENSG00000156194), score: 0.77 PTCH1patched 1 (ENSG00000185920), score: -0.42 PTRH2peptidyl-tRNA hydrolase 2 (ENSG00000141378), score: -0.48 PYCRLpyrroline-5-carboxylate reductase-like (ENSG00000104524), score: 0.47 QSER1glutamine and serine rich 1 (ENSG00000060749), score: 0.48 QSOX1quiescin Q6 sulfhydryl oxidase 1 (ENSG00000116260), score: -0.52 RAC2ras-related C3 botulinum toxin substrate 2 (rho family, small GTP binding protein Rac2) (ENSG00000128340), score: 0.4 RAD52RAD52 homolog (S. cerevisiae) (ENSG00000002016), score: -0.46 RASA3RAS p21 protein activator 3 (ENSG00000185989), score: -0.45 RASL11ARAS-like, family 11, member A (ENSG00000122035), score: -0.43 RASSF6Ras association (RalGDS/AF-6) domain family member 6 (ENSG00000169435), score: 0.43 RCAN1regulator of calcineurin 1 (ENSG00000159200), score: -0.5 RGS18regulator of G-protein signaling 18 (ENSG00000150681), score: 0.48 RIC8Bresistance to inhibitors of cholinesterase 8 homolog B (C. elegans) (ENSG00000111785), score: 0.45 RNF145ring finger protein 145 (ENSG00000145860), score: -0.45 RPS6KA6ribosomal protein S6 kinase, 90kDa, polypeptide 6 (ENSG00000072133), score: 0.44 SDC2syndecan 2 (ENSG00000169439), score: 0.45 SFRS8splicing factor, arginine/serine-rich 8 (suppressor-of-white-apricot homolog, Drosophila) (ENSG00000061936), score: 0.41 SFXN3sideroflexin 3 (ENSG00000107819), score: -0.47 SGSM2small G protein signaling modulator 2 (ENSG00000141258), score: -0.5 SLC15A4solute carrier family 15, member 4 (ENSG00000139370), score: 0.49 SLC16A10solute carrier family 16, member 10 (aromatic amino acid transporter) (ENSG00000112394), score: 0.52 SLC16A12solute carrier family 16, member 12 (monocarboxylic acid transporter 12) (ENSG00000152779), score: 0.41 SLC16A2solute carrier family 16, member 2 (monocarboxylic acid transporter 8) (ENSG00000147100), score: 0.42 SLC25A40solute carrier family 25, member 40 (ENSG00000075303), score: 0.51 SLC38A8solute carrier family 38, member 8 (ENSG00000166558), score: 0.69 SLC46A1solute carrier family 46 (folate transporter), member 1 (ENSG00000076351), score: 0.57 SLC6A6solute carrier family 6 (neurotransmitter transporter, taurine), member 6 (ENSG00000131389), score: 0.42 SMCR7LSmith-Magenis syndrome chromosome region, candidate 7-like (ENSG00000100335), score: 0.4 SNX2sorting nexin 2 (ENSG00000205302), score: 0.58 SPATS2spermatogenesis associated, serine-rich 2 (ENSG00000123352), score: -0.5 STAB2stabilin 2 (ENSG00000136011), score: 0.52 STAMBPSTAM binding protein (ENSG00000124356), score: 0.5 STAP1signal transducing adaptor family member 1 (ENSG00000035720), score: 0.41 STT3BSTT3, subunit of the oligosaccharyltransferase complex, homolog B (S. cerevisiae) (ENSG00000163527), score: 0.59 STX16syntaxin 16 (ENSG00000124222), score: -0.43 TALDO1transaldolase 1 (ENSG00000177156), score: 0.4 TBC1D8BTBC1 domain family, member 8B (with GRAM domain) (ENSG00000133138), score: 0.57 TCF3transcription factor 3 (E2A immunoglobulin enhancer binding factors E12/E47) (ENSG00000071564), score: 0.47 TEX2testis expressed 2 (ENSG00000136478), score: 0.46 TEX264testis expressed 264 (ENSG00000164081), score: 0.49 THYN1thymocyte nuclear protein 1 (ENSG00000151500), score: -0.42 TIAL1TIA1 cytotoxic granule-associated RNA binding protein-like 1 (ENSG00000151923), score: 0.44 TMEM184Ctransmembrane protein 184C (ENSG00000164168), score: 0.73 TMEM41Atransmembrane protein 41A (ENSG00000163900), score: 0.48 TMEM53transmembrane protein 53 (ENSG00000126106), score: 0.46 TMEM9transmembrane protein 9 (ENSG00000116857), score: -0.47 TMPRSS13transmembrane protease, serine 13 (ENSG00000137747), score: 0.41 TNMDtenomodulin (ENSG00000000005), score: 0.47 TOE1target of EGR1, member 1 (nuclear) (ENSG00000132773), score: 0.41 TP53BP1tumor protein p53 binding protein 1 (ENSG00000067369), score: -0.5 TRAF3TNF receptor-associated factor 3 (ENSG00000131323), score: -0.44 TSC22D2TSC22 domain family, member 2 (ENSG00000196428), score: 0.55 TSNtranslin (ENSG00000211460), score: -0.49 TSPAN9tetraspanin 9 (ENSG00000011105), score: 0.43 TTC36tetratricopeptide repeat domain 36 (ENSG00000172425), score: 0.42 TTF2transcription termination factor, RNA polymerase II (ENSG00000116830), score: 0.76 TUBGCP5tubulin, gamma complex associated protein 5 (ENSG00000153575), score: -0.59 TXNDC12thioredoxin domain containing 12 (endoplasmic reticulum) (ENSG00000117862), score: 0.59 TXNRD1thioredoxin reductase 1 (ENSG00000198431), score: 0.4 UBE2G1ubiquitin-conjugating enzyme E2G 1 (UBC7 homolog, yeast) (ENSG00000132388), score: -0.63 UPK3Auroplakin 3A (ENSG00000100373), score: 0.45 UTP20UTP20, small subunit (SSU) processome component, homolog (yeast) (ENSG00000120800), score: 0.43 VPS39vacuolar protein sorting 39 homolog (S. cerevisiae) (ENSG00000166887), score: -0.47 VPS54vacuolar protein sorting 54 homolog (S. cerevisiae) (ENSG00000143952), score: 0.46 WDR37WD repeat domain 37 (ENSG00000047056), score: -0.53 WDR43WD repeat domain 43 (ENSG00000163811), score: 0.47 XCR1chemokine (C motif) receptor 1 (ENSG00000173578), score: 0.45 ZBTB11zinc finger and BTB domain containing 11 (ENSG00000066422), score: -0.44 ZBTB2zinc finger and BTB domain containing 2 (ENSG00000181472), score: 0.4 ZC3H15zinc finger CCCH-type containing 15 (ENSG00000065548), score: -0.62 ZDHHC13zinc finger, DHHC-type containing 13 (ENSG00000177054), score: 0.72 ZNF276zinc finger protein 276 (ENSG00000158805), score: 0.45 ZNF346zinc finger protein 346 (ENSG00000113761), score: -0.54 ZNF512zinc finger protein 512 (ENSG00000243943), score: 0.45 ZPLD1zona pellucida-like domain containing 1 (ENSG00000170044), score: 0.51 ZRANB3zinc finger, RAN-binding domain containing 3 (ENSG00000121988), score: -0.43 ZWILCHZwilch, kinetochore associated, homolog (Drosophila) (ENSG00000174442), score: 0.42
| Id | species | tissue | sex | individual |
|---|---|---|---|---|
| mdo_kd_m_ca1 | mdo | kd | m | _ |
| mdo_kd_f_ca1 | mdo | kd | f | _ |
| mdo_lv_m_ca1 | mdo | lv | m | _ |
| mdo_lv_f_ca1 | mdo | lv | f | _ |