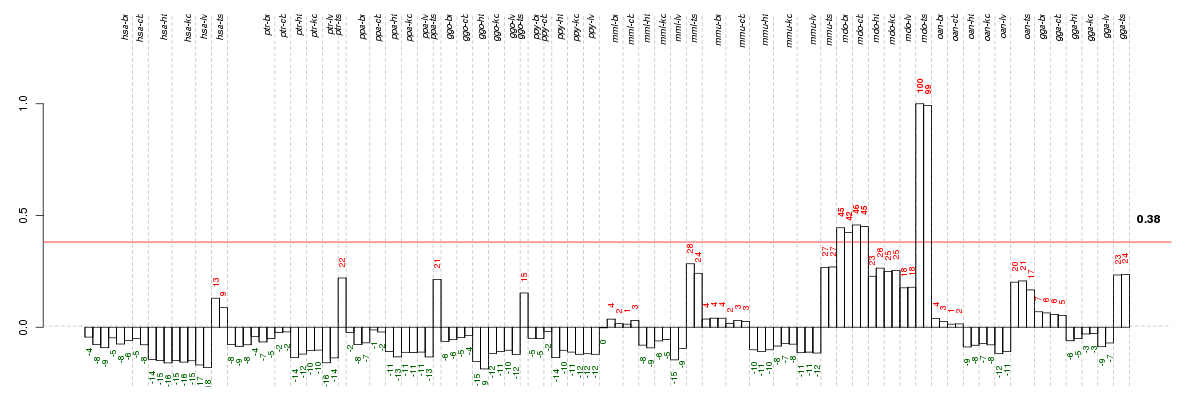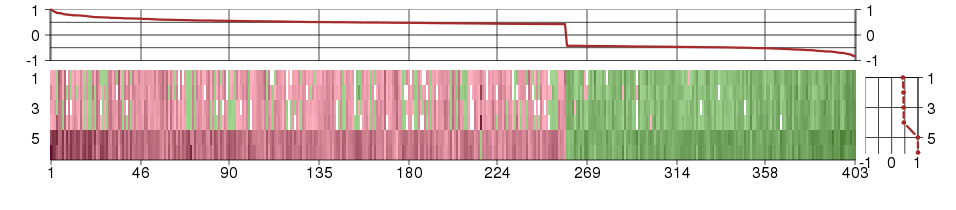

Under-expression is coded with green,
over-expression with red color.

metabolic process
The chemical reactions and pathways, including anabolism and catabolism, by which living organisms transform chemical substances. Metabolic processes typically transform small molecules, but also include macromolecular processes such as DNA repair and replication, and protein synthesis and degradation.
generation of precursor metabolites and energy
The chemical reactions and pathways resulting in the formation of precursor metabolites, substances from which energy is derived, and any process involved in the liberation of energy from these substances.
oxidation reduction
The process of removal or addition of one or more electrons with or without the concomitant removal or addition of a proton or protons.
respiratory electron transport chain
A process whereby a series of electron carriers operate together to transfer electrons from donors such as NADH and FADH2 to any of several different terminal electron acceptors to generate a transmembrane electrochemical gradient.
oxidative phosphorylation
The phosphorylation of ADP to ATP that accompanies the oxidation of a metabolite through the operation of the respiratory chain. Oxidation of compounds establishes a proton gradient across the membrane, providing the energy for ATP synthesis.
mitochondrial electron transport, NADH to ubiquinone
The transfer of electrons from NADH to ubiquinone that occurs during oxidative phosphorylation, mediated by the multisubunit enzyme known as complex I.
electron transport chain
A process whereby a series of electron carriers operate together to transfer electrons from donors to any of several different terminal electron acceptors to generate a transmembrane electrochemical gradient.
nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
Any cellular metabolic process involving nucleobases, nucleosides, nucleotides and nucleic acids.
tRNA metabolic process
The chemical reactions and pathways involving tRNA, transfer RNA, a class of relatively small RNA molecules responsible for mediating the insertion of amino acids into the sequence of nascent polypeptide chains during protein synthesis. Transfer RNA is characterized by the presence of many unusual minor bases, the function of which has not been completely established.
phosphorus metabolic process
The chemical reactions and pathways involving the nonmetallic element phosphorus or compounds that contain phosphorus, usually in the form of a phosphate group (PO4).
phosphate metabolic process
The chemical reactions and pathways involving the phosphate group, the anion or salt of any phosphoric acid.
nitrogen compound metabolic process
The chemical reactions and pathways involving various organic and inorganic nitrogenous compounds; includes nitrogen fixation, nitrification, denitrification, assimilatory/dissimilatory nitrate reduction and the interconversion of nitrogenous organic matter and ammonium.
biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
cellular process
Any process that is carried out at the cellular level, but not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
energy derivation by oxidation of organic compounds
The chemical reactions and pathways by which a cell derives energy from organic compounds; results in the oxidation of the compounds from which energy is released.
RNA metabolic process
The cellular chemical reactions and pathways involving RNA, ribonucleic acid, one of the two main type of nucleic acid, consisting of a long, unbranched macromolecule formed from ribonucleotides joined in 3',5'-phosphodiester linkage.
phosphorylation
The process of introducing a phosphate group into a molecule, usually with the formation of a phosphoric ester, a phosphoric anhydride or a phosphoric amide.
cellular nitrogen compound metabolic process
The chemical reactions and pathways involving various organic and inorganic nitrogenous compounds, as carried out by individual cells.
ncRNA metabolic process
The chemical reactions and pathways involving non-coding RNA transcripts (ncRNAs).
ATP synthesis coupled electron transport
The transfer of electrons through a series of electron donors and acceptors, generating energy that is ultimately used for synthesis of ATP.
mitochondrial ATP synthesis coupled electron transport
The transfer of electrons through a series of electron donors and acceptors, generating energy that is ultimately used for synthesis of ATP, as it occurs in the mitochondrial inner membrane or chloroplast thylakoid membrane.
macromolecule metabolic process
The chemical reactions and pathways involving macromolecules, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass.
cellular metabolic process
The chemical reactions and pathways by which individual cells transform chemical substances.
primary metabolic process
The chemical reactions and pathways involving those compounds which are formed as a part of the normal anabolic and catabolic processes. These processes take place in most, if not all, cells of the organism.
cellular macromolecule metabolic process
The chemical reactions and pathways involving macromolecules, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass, as carried out by individual cells.
cellular respiration
The enzymatic release of energy from organic compounds (especially carbohydrates and fats) which either requires oxygen (aerobic respiration) or does not (anaerobic respiration).
nucleic acid metabolic process
Any cellular metabolic process involving nucleic acids.
all
NA
cellular metabolic process
The chemical reactions and pathways by which individual cells transform chemical substances.
cellular nitrogen compound metabolic process
The chemical reactions and pathways involving various organic and inorganic nitrogenous compounds, as carried out by individual cells.
cellular macromolecule metabolic process
The chemical reactions and pathways involving macromolecules, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass, as carried out by individual cells.
nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
Any cellular metabolic process involving nucleobases, nucleosides, nucleotides and nucleic acids.
electron transport chain
A process whereby a series of electron carriers operate together to transfer electrons from donors to any of several different terminal electron acceptors to generate a transmembrane electrochemical gradient.
ATP synthesis coupled electron transport
The transfer of electrons through a series of electron donors and acceptors, generating energy that is ultimately used for synthesis of ATP.
RNA metabolic process
The cellular chemical reactions and pathways involving RNA, ribonucleic acid, one of the two main type of nucleic acid, consisting of a long, unbranched macromolecule formed from ribonucleotides joined in 3',5'-phosphodiester linkage.
respiratory electron transport chain
A process whereby a series of electron carriers operate together to transfer electrons from donors such as NADH and FADH2 to any of several different terminal electron acceptors to generate a transmembrane electrochemical gradient.
oxidative phosphorylation
The phosphorylation of ADP to ATP that accompanies the oxidation of a metabolite through the operation of the respiratory chain. Oxidation of compounds establishes a proton gradient across the membrane, providing the energy for ATP synthesis.
mitochondrial electron transport, NADH to ubiquinone
The transfer of electrons from NADH to ubiquinone that occurs during oxidative phosphorylation, mediated by the multisubunit enzyme known as complex I.

intracellular
The living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
membrane
Double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
mitochondrion
A semiautonomous, self replicating organelle that occurs in varying numbers, shapes, and sizes in the cytoplasm of virtually all eukaryotic cells. It is notably the site of tissue respiration.
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
cell
The basic structural and functional unit of all organisms. Includes the plasma membrane and any external encapsulating structures such as the cell wall and cell envelope.
cytoplasm
All of the contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures.
mitochondrial envelope
The double lipid bilayer enclosing the mitochondrion and separating its contents from the cell cytoplasm; includes the intermembrane space.
mitochondrial inner membrane
The inner, i.e. lumen-facing, lipid bilayer of the mitochondrial envelope. It is highly folded to form cristae.
mitochondrial respiratory chain
The protein complexes that form the mitochondrial electron transport system (the respiratory chain), associated with the inner mitochondrial membrane. The respiratory chain complexes transfer electrons from an electron donor to an electron acceptor and are associated with a proton pump to create a transmembrane electrochemical gradient.
mitochondrial respiratory chain complex I
A protein complex located in the mitochondrial inner membrane that forms part of the mitochondrial respiratory chain. It contains about 25 different polypeptide subunits, including NADH dehydrogenase (ubiquinone), flavin mononucleotide and several different iron-sulfur clusters containing non-heme iron. The iron undergoes oxidation-reduction between Fe(II) and Fe(III), and catalyzes proton translocation linked to the oxidation of NADH by ubiquinone.
organelle inner membrane
The inner, i.e. lumen-facing, lipid bilayer of an organelle envelope; usually highly selective to most ions and metabolites.
NADH dehydrogenase complex
An integral membrane complex that possesses NADH oxidoreductase activity. The complex is one of the components of the electron transport chain. It catalyzes the transfer of a pair of electrons from NADH to a quinone.
organelle membrane
The lipid bilayer surrounding an organelle.
mitochondrial membrane
Either of the lipid bilayers that surround the mitochondrion and form the mitochondrial envelope.
organelle envelope
A double membrane structure enclosing an organelle, including two lipid bilayers and the region between them. In some cases, an organelle envelope may have more than two membranes.
envelope
A multilayered structure surrounding all or part of a cell; encompasses one or more lipid bilayers, and may include a cell wall layer; also includes the space between layers.
macromolecular complex
A stable assembly of two or more macromolecules, i.e. proteins, nucleic acids, carbohydrates or lipids, in which the constituent parts function together.
organelle
Organized structure of distinctive morphology and function. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton, and prokaryotic structures such as anammoxosomes and pirellulosomes. Excludes the plasma membrane.
membrane-bounded organelle
Organized structure of distinctive morphology and function, bounded by a single or double lipid bilayer membrane. Includes the nucleus, mitochondria, plastids, vacuoles, and vesicles. Excludes the plasma membrane.
intracellular organelle
Organized structure of distinctive morphology and function, occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton. Excludes the plasma membrane.
intracellular membrane-bounded organelle
Organized structure of distinctive morphology and function, bounded by a single or double lipid bilayer membrane and occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, and vesicles. Excludes the plasma membrane.
protein complex
Any macromolecular complex composed of two or more polypeptide subunits, which may or may not be identical. Protein complexes may have other associated non-protein prosthetic groups, such as nucleotides, metal ions or other small molecules.
organelle part
Any constituent part of an organelle, an organized structure of distinctive morphology and function. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton, but excludes the plasma membrane.
intracellular part
Any constituent part of the living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
membrane part
Any constituent part of a membrane, a double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
mitochondrial part
Any constituent part of a mitochondrion, a semiautonomous, self replicating organelle that occurs in varying numbers, shapes, and sizes in the cytoplasm of virtually all eukaryotic cells. It is notably the site of tissue respiration.
cytoplasmic part
Any constituent part of the cytoplasm, all of the contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures.
intracellular organelle part
A constituent part of an intracellular organelle, an organized structure of distinctive morphology and function, occurring within the cell. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton but excludes the plasma membrane.
mitochondrial membrane part
Any constituent part of a mitochondrial membrane, either of the lipid bilayers that surround the mitochondrion and form the mitochondrial envelope.
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
respiratory chain complex I
Respiratory chain complex I is an enzyme of the respiratory chain. It consists of at least 34 polypeptide chains and is L-shaped, with a horizontal arm lying in the membrane and a vertical arm that projects into the matrix. The electrons of NADH enter the chain at this complex.
respiratory chain
The protein complexes that form the electron transport system (the respiratory chain), associated with a cell membrane, usually the plasma membrane (in prokaryotes) or the inner mitochondrial membrane (on eukaryotes). The respiratory chain complexes transfer electrons from an electron donor to an electron acceptor and are associated with a proton pump to create a transmembrane electrochemical gradient.
all
NA
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
organelle part
Any constituent part of an organelle, an organized structure of distinctive morphology and function. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton, but excludes the plasma membrane.
organelle membrane
The lipid bilayer surrounding an organelle.
intracellular membrane-bounded organelle
Organized structure of distinctive morphology and function, bounded by a single or double lipid bilayer membrane and occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, and vesicles. Excludes the plasma membrane.
intracellular organelle part
A constituent part of an intracellular organelle, an organized structure of distinctive morphology and function, occurring within the cell. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton but excludes the plasma membrane.
organelle envelope
A double membrane structure enclosing an organelle, including two lipid bilayers and the region between them. In some cases, an organelle envelope may have more than two membranes.
intracellular part
Any constituent part of the living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
organelle membrane
The lipid bilayer surrounding an organelle.
membrane part
Any constituent part of a membrane, a double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
organelle envelope
A double membrane structure enclosing an organelle, including two lipid bilayers and the region between them. In some cases, an organelle envelope may have more than two membranes.
organelle inner membrane
The inner, i.e. lumen-facing, lipid bilayer of an organelle envelope; usually highly selective to most ions and metabolites.
NADH dehydrogenase complex
An integral membrane complex that possesses NADH oxidoreductase activity. The complex is one of the components of the electron transport chain. It catalyzes the transfer of a pair of electrons from NADH to a quinone.
intracellular organelle
Organized structure of distinctive morphology and function, occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton. Excludes the plasma membrane.
intracellular organelle part
A constituent part of an intracellular organelle, an organized structure of distinctive morphology and function, occurring within the cell. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton but excludes the plasma membrane.
NADH dehydrogenase complex
An integral membrane complex that possesses NADH oxidoreductase activity. The complex is one of the components of the electron transport chain. It catalyzes the transfer of a pair of electrons from NADH to a quinone.
organelle inner membrane
The inner, i.e. lumen-facing, lipid bilayer of an organelle envelope; usually highly selective to most ions and metabolites.
mitochondrial inner membrane
The inner, i.e. lumen-facing, lipid bilayer of the mitochondrial envelope. It is highly folded to form cristae.
mitochondrial membrane part
Any constituent part of a mitochondrial membrane, either of the lipid bilayers that surround the mitochondrion and form the mitochondrial envelope.
mitochondrial envelope
The double lipid bilayer enclosing the mitochondrion and separating its contents from the cell cytoplasm; includes the intermembrane space.
mitochondrial membrane
Either of the lipid bilayers that surround the mitochondrion and form the mitochondrial envelope.
mitochondrial membrane part
Any constituent part of a mitochondrial membrane, either of the lipid bilayers that surround the mitochondrion and form the mitochondrial envelope.
cytoplasmic part
Any constituent part of the cytoplasm, all of the contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures.
mitochondrion
A semiautonomous, self replicating organelle that occurs in varying numbers, shapes, and sizes in the cytoplasm of virtually all eukaryotic cells. It is notably the site of tissue respiration.
mitochondrial part
Any constituent part of a mitochondrion, a semiautonomous, self replicating organelle that occurs in varying numbers, shapes, and sizes in the cytoplasm of virtually all eukaryotic cells. It is notably the site of tissue respiration.
mitochondrial respiratory chain
The protein complexes that form the mitochondrial electron transport system (the respiratory chain), associated with the inner mitochondrial membrane. The respiratory chain complexes transfer electrons from an electron donor to an electron acceptor and are associated with a proton pump to create a transmembrane electrochemical gradient.
respiratory chain complex I
Respiratory chain complex I is an enzyme of the respiratory chain. It consists of at least 34 polypeptide chains and is L-shaped, with a horizontal arm lying in the membrane and a vertical arm that projects into the matrix. The electrons of NADH enter the chain at this complex.
mitochondrial respiratory chain complex I
A protein complex located in the mitochondrial inner membrane that forms part of the mitochondrial respiratory chain. It contains about 25 different polypeptide subunits, including NADH dehydrogenase (ubiquinone), flavin mononucleotide and several different iron-sulfur clusters containing non-heme iron. The iron undergoes oxidation-reduction between Fe(II) and Fe(III), and catalyzes proton translocation linked to the oxidation of NADH by ubiquinone.
mitochondrial membrane
Either of the lipid bilayers that surround the mitochondrion and form the mitochondrial envelope.
mitochondrial part
Any constituent part of a mitochondrion, a semiautonomous, self replicating organelle that occurs in varying numbers, shapes, and sizes in the cytoplasm of virtually all eukaryotic cells. It is notably the site of tissue respiration.
mitochondrial respiratory chain
The protein complexes that form the mitochondrial electron transport system (the respiratory chain), associated with the inner mitochondrial membrane. The respiratory chain complexes transfer electrons from an electron donor to an electron acceptor and are associated with a proton pump to create a transmembrane electrochemical gradient.
mitochondrial respiratory chain complex I
A protein complex located in the mitochondrial inner membrane that forms part of the mitochondrial respiratory chain. It contains about 25 different polypeptide subunits, including NADH dehydrogenase (ubiquinone), flavin mononucleotide and several different iron-sulfur clusters containing non-heme iron. The iron undergoes oxidation-reduction between Fe(II) and Fe(III), and catalyzes proton translocation linked to the oxidation of NADH by ubiquinone.

magnesium ion binding
Interacting selectively and non-covalently with magnesium (Mg) ions.
molecular_function
Elemental activities, such as catalysis or binding, describing the actions of a gene product at the molecular level. A given gene product may exhibit one or more molecular functions.
nucleic acid binding
Interacting selectively and non-covalently with any nucleic acid.
DNA binding
Any molecular function by which a gene product interacts selectively with DNA (deoxyribonucleic acid).
damaged DNA binding
Interacting selectively and non-covalently with damaged DNA.
catalytic activity
Catalysis of a biochemical reaction at physiological temperatures. In biologically catalyzed reactions, the reactants are known as substrates, and the catalysts are naturally occurring macromolecular substances known as enzymes. Enzymes possess specific binding sites for substrates, and are usually composed wholly or largely of protein, but RNA that has catalytic activity (ribozyme) is often also regarded as enzymatic.
NADH dehydrogenase activity
Catalysis of the reaction: NADH + H+ + acceptor = NAD+ + reduced acceptor.
binding
The selective, non-covalent, often stoichiometric, interaction of a molecule with one or more specific sites on another molecule.
metal ion binding
Interacting selectively and non-covalently with any metal ion.
oxidoreductase activity
Catalysis of an oxidation-reduction (redox) reaction, a reversible chemical reaction in which the oxidation state of an atom or atoms within a molecule is altered. One substrate acts as a hydrogen or electron donor and becomes oxidized, while the other acts as hydrogen or electron acceptor and becomes reduced.
NADH dehydrogenase (ubiquinone) activity
Catalysis of the reaction: NADH + H+ + ubiquinone = NAD+ + ubiquinol.
oxidoreductase activity, acting on NADH or NADPH
Catalysis of an oxidation-reduction (redox) reaction in which NADH or NADPH acts as a hydrogen or electron donor and reduces a hydrogen or electron acceptor.
oxidoreductase activity, acting on NADH or NADPH, quinone or similar compound as acceptor
Catalysis of an oxidation-reduction (redox) reaction in which NADH or NADPH acts as a hydrogen or electron donor and reduces a quinone or a similar acceptor molecule.
ion binding
Interacting selectively and non-covalently with ions, charged atoms or groups of atoms.
cation binding
Interacting selectively and non-covalently with cations, charged atoms or groups of atoms with a net positive charge.
NADH dehydrogenase (quinone) activity
Catalysis of the reaction: NADH + H+ + a quinone = NAD+ + a quinol.
all
NA
NADH dehydrogenase (quinone) activity
Catalysis of the reaction: NADH + H+ + a quinone = NAD+ + a quinol.
| Id | Pvalue | ExpCount | Count | Size | Term |
|---|---|---|---|---|---|
| 05012 | 4.787e-06 | 1.71 | 12 | 27 | Parkinson's disease |
| 00190 | 7.740e-04 | 1.9 | 10 | 30 | Oxidative phosphorylation |
AASDHPPTaminoadipate-semialdehyde dehydrogenase-phosphopantetheinyl transferase (ENSG00000149313), score: -0.52 ABCB7ATP-binding cassette, sub-family B (MDR/TAP), member 7 (ENSG00000131269), score: -0.44 ACAP2ArfGAP with coiled-coil, ankyrin repeat and PH domains 2 (ENSG00000114331), score: 0.63 ACER1alkaline ceramidase 1 (ENSG00000167769), score: 0.69 ACVR1activin A receptor, type I (ENSG00000115170), score: -0.48 ADCarginine decarboxylase (ENSG00000142920), score: 0.49 ADCY7adenylate cyclase 7 (ENSG00000121281), score: -0.47 ADSLadenylosuccinate lyase (ENSG00000100354), score: 0.46 AGBL2ATP/GTP binding protein-like 2 (ENSG00000165923), score: 0.55 AHSA2AHA1, activator of heat shock 90kDa protein ATPase homolog 2 (yeast) (ENSG00000173209), score: -0.66 AIREautoimmune regulator (ENSG00000160224), score: 0.79 ALG2asparagine-linked glycosylation 2, alpha-1,3-mannosyltransferase homolog (S. cerevisiae) (ENSG00000119523), score: -0.47 ALKBH2alkB, alkylation repair homolog 2 (E. coli) (ENSG00000189046), score: 0.46 ANKRD13Cankyrin repeat domain 13C (ENSG00000118454), score: 0.43 ANKRD16ankyrin repeat domain 16 (ENSG00000134461), score: -0.47 ANXA6annexin A6 (ENSG00000197043), score: -0.54 APAF1apoptotic peptidase activating factor 1 (ENSG00000120868), score: 0.59 APTXaprataxin (ENSG00000137074), score: 0.69 ARHGAP22Rho GTPase activating protein 22 (ENSG00000128805), score: 0.55 ARL9ADP-ribosylation factor-like 9 (ENSG00000196503), score: 0.45 ARSKarylsulfatase family, member K (ENSG00000164291), score: 0.47 ASH2Lash2 (absent, small, or homeotic)-like (Drosophila) (ENSG00000129691), score: 0.43 ATG4CATG4 autophagy related 4 homolog C (S. cerevisiae) (ENSG00000125703), score: 0.54 ATP2C2ATPase, Ca++ transporting, type 2C, member 2 (ENSG00000064270), score: 0.65 B3GALTLbeta 1,3-galactosyltransferase-like (ENSG00000187676), score: -0.44 BARX2BARX homeobox 2 (ENSG00000043039), score: 0.44 BBS7Bardet-Biedl syndrome 7 (ENSG00000138686), score: 0.51 BCL2L13BCL2-like 13 (apoptosis facilitator) (ENSG00000099968), score: -0.59 BLVRAbiliverdin reductase A (ENSG00000106605), score: -0.66 BPGM2,3-bisphosphoglycerate mutase (ENSG00000172331), score: 0.49 BRPF1bromodomain and PHD finger containing, 1 (ENSG00000156983), score: 0.57 C10orf137chromosome 10 open reading frame 137 (ENSG00000107938), score: 0.44 C10orf28chromosome 10 open reading frame 28 (ENSG00000166024), score: 0.59 C11orf46chromosome 11 open reading frame 46 (ENSG00000152219), score: 0.86 C11orf57chromosome 11 open reading frame 57 (ENSG00000150776), score: -0.53 C12orf23chromosome 12 open reading frame 23 (ENSG00000151135), score: 0.57 C12orf48chromosome 12 open reading frame 48 (ENSG00000185480), score: 0.43 C12orf50chromosome 12 open reading frame 50 (ENSG00000165805), score: 0.44 C12orf63chromosome 12 open reading frame 63 (ENSG00000188596), score: 0.5 C13orf23chromosome 13 open reading frame 23 (ENSG00000120685), score: 0.46 C13orf31chromosome 13 open reading frame 31 (ENSG00000179630), score: 0.67 C14orf118chromosome 14 open reading frame 118 (ENSG00000089916), score: 0.6 C14orf159chromosome 14 open reading frame 159 (ENSG00000133943), score: -0.46 C17orf71chromosome 17 open reading frame 71 (ENSG00000167447), score: 0.62 C18orf55chromosome 18 open reading frame 55 (ENSG00000075336), score: -0.63 C1orf130chromosome 1 open reading frame 130 (ENSG00000184454), score: 0.47 C1orf151chromosome 1 open reading frame 151 (ENSG00000173436), score: -0.49 C1orf159chromosome 1 open reading frame 159 (ENSG00000131591), score: 0.44 C1orf9chromosome 1 open reading frame 9 (ENSG00000094975), score: 0.54 C2CD2C2 calcium-dependent domain containing 2 (ENSG00000157617), score: -0.51 C3orf39chromosome 3 open reading frame 39 (ENSG00000144647), score: 0.67 C5orf24chromosome 5 open reading frame 24 (ENSG00000181904), score: -0.48 C5orf30chromosome 5 open reading frame 30 (ENSG00000181751), score: 0.5 C6orf145chromosome 6 open reading frame 145 (ENSG00000168994), score: -0.43 C6orf182chromosome 6 open reading frame 182 (ENSG00000183137), score: 0.45 C6orf204chromosome 6 open reading frame 204 (ENSG00000111860), score: 0.43 C6orf89chromosome 6 open reading frame 89 (ENSG00000198663), score: 0.56 C9orf103chromosome 9 open reading frame 103 (ENSG00000148057), score: -0.49 CA13carbonic anhydrase XIII (ENSG00000185015), score: 0.51 CARS2cysteinyl-tRNA synthetase 2, mitochondrial (putative) (ENSG00000134905), score: -0.5 CCDC135coiled-coil domain containing 135 (ENSG00000159625), score: 0.47 CCDC3coiled-coil domain containing 3 (ENSG00000151468), score: -0.44 CCDC37coiled-coil domain containing 37 (ENSG00000163885), score: 0.52 CCDC43coiled-coil domain containing 43 (ENSG00000180329), score: 0.69 CCDC67coiled-coil domain containing 67 (ENSG00000165325), score: 0.43 CDC42BPBCDC42 binding protein kinase beta (DMPK-like) (ENSG00000198752), score: -0.63 CDCA3cell division cycle associated 3 (ENSG00000111665), score: 0.58 CDK10cyclin-dependent kinase 10 (ENSG00000185324), score: -0.44 CDKL5cyclin-dependent kinase-like 5 (ENSG00000008086), score: 0.49 CDKN1Bcyclin-dependent kinase inhibitor 1B (p27, Kip1) (ENSG00000111276), score: -0.44 CDR2Lcerebellar degeneration-related protein 2-like (ENSG00000109089), score: 0.43 CEP152centrosomal protein 152kDa (ENSG00000103995), score: 0.49 CFDP1craniofacial development protein 1 (ENSG00000153774), score: -0.48 CHMP6chromatin modifying protein 6 (ENSG00000176108), score: -0.48 CISD2CDGSH iron sulfur domain 2 (ENSG00000145354), score: -0.49 CKAP4cytoskeleton-associated protein 4 (ENSG00000136026), score: -0.47 CNOT2CCR4-NOT transcription complex, subunit 2 (ENSG00000111596), score: 0.51 CNOT8CCR4-NOT transcription complex, subunit 8 (ENSG00000155508), score: 0.61 CNTFciliary neurotrophic factor (ENSG00000186660), score: 0.6 COX1cytochrome c oxidase subunit I (ENSG00000198804), score: -0.83 COX4I1cytochrome c oxidase subunit IV isoform 1 (ENSG00000131143), score: -0.61 CPEB2cytoplasmic polyadenylation element binding protein 2 (ENSG00000137449), score: 0.48 CPMcarboxypeptidase M (ENSG00000135678), score: -0.42 CPZcarboxypeptidase Z (ENSG00000109625), score: 0.53 CRTAPcartilage associated protein (ENSG00000170275), score: -0.7 CSRP2BPCSRP2 binding protein (ENSG00000149474), score: -0.47 CYTBcytochrome b (ENSG00000198727), score: -0.81 CYTH1cytohesin 1 (ENSG00000108669), score: -0.45 CYTSBcytospin B (ENSG00000128487), score: 0.45 DCP1BDCP1 decapping enzyme homolog B (S. cerevisiae) (ENSG00000151065), score: 0.43 DCTN5dynactin 5 (p25) (ENSG00000166847), score: -0.52 DDX1DEAD (Asp-Glu-Ala-Asp) box polypeptide 1 (ENSG00000079785), score: 0.65 DDX41DEAD (Asp-Glu-Ala-Asp) box polypeptide 41 (ENSG00000183258), score: -0.59 DENND2CDENN/MADD domain containing 2C (ENSG00000175984), score: 0.53 DGUOKdeoxyguanosine kinase (ENSG00000114956), score: -0.72 DHX30DEAH (Asp-Glu-Ala-His) box polypeptide 30 (ENSG00000132153), score: 0.48 DNM1Ldynamin 1-like (ENSG00000087470), score: 0.47 DUS4Ldihydrouridine synthase 4-like (S. cerevisiae) (ENSG00000105865), score: -0.46 DUSP4dual specificity phosphatase 4 (ENSG00000120875), score: 0.65 E2F8E2F transcription factor 8 (ENSG00000129173), score: 0.48 ECDecdysoneless homolog (Drosophila) (ENSG00000122882), score: 0.95 EFCAB7EF-hand calcium binding domain 7 (ENSG00000203965), score: 0.5 EIF2B1eukaryotic translation initiation factor 2B, subunit 1 alpha, 26kDa (ENSG00000111361), score: -0.45 EIF2C2eukaryotic translation initiation factor 2C, 2 (ENSG00000123908), score: -0.51 EIF3Ieukaryotic translation initiation factor 3, subunit I (ENSG00000084623), score: -0.53 ENO4enolase family member 4 (ENSG00000188316), score: 0.49 EPS15epidermal growth factor receptor pathway substrate 15 (ENSG00000085832), score: 0.53 ERGIC1endoplasmic reticulum-golgi intermediate compartment (ERGIC) 1 (ENSG00000113719), score: -0.48 EXOC3exocyst complex component 3 (ENSG00000180104), score: -0.47 FAF2Fas associated factor family member 2 (ENSG00000113194), score: 0.47 FAM135Bfamily with sequence similarity 135, member B (ENSG00000147724), score: 0.46 FAM13Cfamily with sequence similarity 13, member C (ENSG00000148541), score: 0.5 FAM161Afamily with sequence similarity 161, member A (ENSG00000170264), score: 0.52 FAM172Afamily with sequence similarity 172, member A (ENSG00000113391), score: 0.45 FAM179Afamily with sequence similarity 179, member A (ENSG00000189350), score: 0.55 FAM18Afamily with sequence similarity 18, member A (ENSG00000166676), score: 0.49 FAM20Bfamily with sequence similarity 20, member B (ENSG00000116199), score: 0.43 FAM54Afamily with sequence similarity 54, member A (ENSG00000146410), score: 0.55 FAM59Afamily with sequence similarity 59, member A (ENSG00000141441), score: -0.42 FAM73Afamily with sequence similarity 73, member A (ENSG00000180488), score: 0.5 FARP2FERM, RhoGEF and pleckstrin domain protein 2 (ENSG00000006607), score: 0.45 FARS2phenylalanyl-tRNA synthetase 2, mitochondrial (ENSG00000145982), score: -0.45 FBXO15F-box protein 15 (ENSG00000141665), score: 0.57 FBXO22F-box protein 22 (ENSG00000167196), score: 0.59 FBXO3F-box protein 3 (ENSG00000110429), score: 0.44 FESfeline sarcoma oncogene (ENSG00000182511), score: 0.49 FEZ2fasciculation and elongation protein zeta 2 (zygin II) (ENSG00000171055), score: -0.45 FGF10fibroblast growth factor 10 (ENSG00000070193), score: 0.58 FGF4fibroblast growth factor 4 (ENSG00000075388), score: 0.58 FGL2fibrinogen-like 2 (ENSG00000127951), score: -0.53 FKBP14FK506 binding protein 14, 22 kDa (ENSG00000106080), score: 0.69 FKBP15FK506 binding protein 15, 133kDa (ENSG00000119321), score: 0.55 FKTNfukutin (ENSG00000106692), score: 0.46 FLVCR1feline leukemia virus subgroup C cellular receptor 1 (ENSG00000162769), score: 0.49 FN3KRPfructosamine 3 kinase related protein (ENSG00000141560), score: -0.42 FOXN1forkhead box N1 (ENSG00000109101), score: 1 FUT10fucosyltransferase 10 (alpha (1,3) fucosyltransferase) (ENSG00000172728), score: 0.76 GABREgamma-aminobutyric acid (GABA) A receptor, epsilon (ENSG00000102287), score: 0.64 GAPVD1GTPase activating protein and VPS9 domains 1 (ENSG00000165219), score: 0.43 GARSglycyl-tRNA synthetase (ENSG00000106105), score: 0.63 GDPD4glycerophosphodiester phosphodiesterase domain containing 4 (ENSG00000178795), score: 0.72 GEMC1geminin coiled-coil domain-containing protein 1 (ENSG00000205835), score: 0.71 GEMIN5gem (nuclear organelle) associated protein 5 (ENSG00000082516), score: 0.67 GGA1golgi-associated, gamma adaptin ear containing, ARF binding protein 1 (ENSG00000100083), score: 0.48 GK5glycerol kinase 5 (putative) (ENSG00000175066), score: 0.46 GLB1galactosidase, beta 1 (ENSG00000170266), score: -0.7 GLI3GLI family zinc finger 3 (ENSG00000106571), score: 0.57 GLMNglomulin, FKBP associated protein (ENSG00000174842), score: 0.65 GM2AGM2 ganglioside activator (ENSG00000196743), score: -0.5 GNPTGN-acetylglucosamine-1-phosphate transferase, gamma subunit (ENSG00000090581), score: -0.43 GORASP2golgi reassembly stacking protein 2, 55kDa (ENSG00000115806), score: -0.43 GPR1G protein-coupled receptor 1 (ENSG00000183671), score: 0.57 GPR107G protein-coupled receptor 107 (ENSG00000148358), score: -0.46 GRAMD3GRAM domain containing 3 (ENSG00000155324), score: -0.58 GRB10growth factor receptor-bound protein 10 (ENSG00000106070), score: -0.56 GRXCR1glutaredoxin, cysteine rich 1 (ENSG00000215203), score: 0.56 GTPBP2GTP binding protein 2 (ENSG00000172432), score: 0.46 HAPLN1hyaluronan and proteoglycan link protein 1 (ENSG00000145681), score: 0.43 HEATR6HEAT repeat containing 6 (ENSG00000068097), score: 0.77 HEBP1heme binding protein 1 (ENSG00000013583), score: -0.43 HELLShelicase, lymphoid-specific (ENSG00000119969), score: 0.54 HESX1HESX homeobox 1 (ENSG00000163666), score: 0.44 HNRNPUheterogeneous nuclear ribonucleoprotein U (scaffold attachment factor A) (ENSG00000153187), score: 0.59 HORMAD2HORMA domain containing 2 (ENSG00000176635), score: 0.43 HOXB13homeobox B13 (ENSG00000159184), score: 0.87 HPRT1hypoxanthine phosphoribosyltransferase 1 (ENSG00000165704), score: 0.44 HYIhydroxypyruvate isomerase (putative) (ENSG00000178922), score: -0.55 IFNAR1interferon (alpha, beta and omega) receptor 1 (ENSG00000142166), score: -0.52 IFT74intraflagellar transport 74 homolog (Chlamydomonas) (ENSG00000096872), score: 0.44 IFT81intraflagellar transport 81 homolog (Chlamydomonas) (ENSG00000122970), score: 0.59 IKIK cytokine, down-regulator of HLA II (ENSG00000113141), score: -0.47 IKBIPIKBKB interacting protein (ENSG00000166130), score: 0.6 IKBKBinhibitor of kappa light polypeptide gene enhancer in B-cells, kinase beta (ENSG00000104365), score: -0.7 IL10RBinterleukin 10 receptor, beta (ENSG00000243646), score: -0.52 INPP4Binositol polyphosphate-4-phosphatase, type II, 105kDa (ENSG00000109452), score: 0.57 INVSinversin (ENSG00000119509), score: -0.56 IPPintracisternal A particle-promoted polypeptide (ENSG00000197429), score: 0.81 IRAK1BP1interleukin-1 receptor-associated kinase 1 binding protein 1 (ENSG00000146243), score: 0.51 IWS1IWS1 homolog (S. cerevisiae) (ENSG00000163166), score: 0.52 JMJD6jumonji domain containing 6 (ENSG00000070495), score: -0.44 KATNB1katanin p80 (WD repeat containing) subunit B 1 (ENSG00000140854), score: 0.44 KBTBD8kelch repeat and BTB (POZ) domain containing 8 (ENSG00000163376), score: 0.57 KCTD20potassium channel tetramerisation domain containing 20 (ENSG00000112078), score: 0.43 KDM1Blysine (K)-specific demethylase 1B (ENSG00000165097), score: -0.47 KDM3Alysine (K)-specific demethylase 3A (ENSG00000115548), score: 0.62 KIAA0100KIAA0100 (ENSG00000007202), score: -0.48 KIAA0146KIAA0146 (ENSG00000164808), score: -0.45 KIAA0174KIAA0174 (ENSG00000182149), score: -0.58 KIAA0895KIAA0895 (ENSG00000164542), score: 0.53 KIAA1609KIAA1609 (ENSG00000140950), score: 0.82 KIAA1712KIAA1712 (ENSG00000164118), score: 0.54 KIF14kinesin family member 14 (ENSG00000118193), score: 0.55 KIF24kinesin family member 24 (ENSG00000186638), score: 0.52 KIF27kinesin family member 27 (ENSG00000165115), score: 0.43 LAMC3laminin, gamma 3 (ENSG00000050555), score: 0.55 LANCL2LanC lantibiotic synthetase component C-like 2 (bacterial) (ENSG00000132434), score: 0.54 LARP4BLa ribonucleoprotein domain family, member 4B (ENSG00000107929), score: -0.52 LCA5Leber congenital amaurosis 5 (ENSG00000135338), score: 0.61 LCTlactase (ENSG00000115850), score: 0.44 LEMD2LEM domain containing 2 (ENSG00000161904), score: -0.43 LEPRE1leucine proline-enriched proteoglycan (leprecan) 1 (ENSG00000117385), score: 0.47 LIMD2LIM domain containing 2 (ENSG00000136490), score: 0.49 LOC100133760similar to Jumonji, AT rich interactive domain 1B (RBP2-like) (ENSG00000117139), score: 0.47 LOC100134291similar to mitogen-activated protein kinase phosphatase x (ENSG00000112679), score: -0.43 LOC100290337similar to damage-specific DNA binding protein 1 (ENSG00000167986), score: 0.44 LONRF3LON peptidase N-terminal domain and ring finger 3 (ENSG00000175556), score: 0.56 LRATlecithin retinol acyltransferase (phosphatidylcholine--retinol O-acyltransferase) (ENSG00000121207), score: 0.55 LSM10LSM10, U7 small nuclear RNA associated (ENSG00000181817), score: -0.48 LTA4Hleukotriene A4 hydrolase (ENSG00000111144), score: -0.5 MAKmale germ cell-associated kinase (ENSG00000111837), score: 0.5 MAMLD1mastermind-like domain containing 1 (ENSG00000013619), score: 0.43 MAP3K4mitogen-activated protein kinase kinase kinase 4 (ENSG00000085511), score: -0.48 MAPK12mitogen-activated protein kinase 12 (ENSG00000188130), score: -0.45 MAPK15mitogen-activated protein kinase 15 (ENSG00000181085), score: 0.45 MAPKAPK3mitogen-activated protein kinase-activated protein kinase 3 (ENSG00000114738), score: -0.47 MECRmitochondrial trans-2-enoyl-CoA reductase (ENSG00000116353), score: -0.45 MED1mediator complex subunit 1 (ENSG00000125686), score: 0.51 MED22mediator complex subunit 22 (ENSG00000148297), score: -0.46 MEMO1mediator of cell motility 1 (ENSG00000162961), score: -0.63 METRNLmeteorin, glial cell differentiation regulator-like (ENSG00000176845), score: -0.48 MID2midline 2 (ENSG00000080561), score: 0.45 MKS1Meckel syndrome, type 1 (ENSG00000011143), score: 0.54 MPLmyeloproliferative leukemia virus oncogene (ENSG00000117400), score: 0.45 MPP6membrane protein, palmitoylated 6 (MAGUK p55 subfamily member 6) (ENSG00000105926), score: 0.43 MRPL2mitochondrial ribosomal protein L2 (ENSG00000112651), score: -0.46 MRPL44mitochondrial ribosomal protein L44 (ENSG00000135900), score: -0.48 MRPS16mitochondrial ribosomal protein S16 (ENSG00000182180), score: -0.42 MSH6mutS homolog 6 (E. coli) (ENSG00000116062), score: 0.49 MTF2metal response element binding transcription factor 2 (ENSG00000143033), score: 0.46 MTIF3mitochondrial translational initiation factor 3 (ENSG00000122033), score: -0.64 MTMR3myotubularin related protein 3 (ENSG00000100330), score: 0.51 MTRR5-methyltetrahydrofolate-homocysteine methyltransferase reductase (ENSG00000124275), score: -0.42 MYBv-myb myeloblastosis viral oncogene homolog (avian) (ENSG00000118513), score: 0.75 MYL1myosin, light chain 1, alkali; skeletal, fast (ENSG00000168530), score: 0.58 N4BP1NEDD4 binding protein 1 (ENSG00000102921), score: 0.53 NARFnuclear prelamin A recognition factor (ENSG00000141562), score: -0.65 NAT9N-acetyltransferase 9 (GCN5-related, putative) (ENSG00000109065), score: -0.54 ND2MTND2 (ENSG00000198763), score: -0.75 ND4NADH dehydrogenase, subunit 4 (complex I) (ENSG00000198886), score: -0.5 ND6NADH dehydrogenase, subunit 6 (complex I) (ENSG00000198695), score: -0.57 NDUFA8NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 8, 19kDa (ENSG00000119421), score: -0.52 NDUFB10NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 10, 22kDa (ENSG00000140990), score: -0.47 NDUFB9NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 9, 22kDa (ENSG00000147684), score: -0.49 NEIL1nei endonuclease VIII-like 1 (E. coli) (ENSG00000140398), score: -0.46 NEK11NIMA (never in mitosis gene a)- related kinase 11 (ENSG00000114670), score: 0.48 NFKBIAnuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha (ENSG00000100906), score: -0.46 NIF3L1NIF3 NGG1 interacting factor 3-like 1 (S. pombe) (ENSG00000196290), score: 0.49 NOXO1NADPH oxidase organizer 1 (ENSG00000196408), score: 0.6 NPSR1neuropeptide S receptor 1 (ENSG00000187258), score: 0.7 NSUN2NOP2/Sun domain family, member 2 (ENSG00000037474), score: -0.57 NTSneurotensin (ENSG00000133636), score: 0.57 NUDCD3NudC domain containing 3 (ENSG00000015676), score: -0.5 NUDT3nudix (nucleoside diphosphate linked moiety X)-type motif 3 (ENSG00000112664), score: -0.45 NUF2NUF2, NDC80 kinetochore complex component, homolog (S. cerevisiae) (ENSG00000143228), score: 0.5 NUP153nucleoporin 153kDa (ENSG00000124789), score: 0.63 NUP54nucleoporin 54kDa (ENSG00000138750), score: 0.57 NUP85nucleoporin 85kDa (ENSG00000125450), score: 0.43 OGFOD12-oxoglutarate and iron-dependent oxygenase domain containing 1 (ENSG00000087263), score: -0.42 OSTF1osteoclast stimulating factor 1 (ENSG00000134996), score: -0.5 OTOL1otolin 1 homolog (zebrafish) (ENSG00000182447), score: 0.45 PABPC1Lpoly(A) binding protein, cytoplasmic 1-like (ENSG00000101104), score: 0.47 PAPD4PAP associated domain containing 4 (ENSG00000164329), score: 0.64 PARK7Parkinson disease (autosomal recessive, early onset) 7 (ENSG00000116288), score: -0.6 PDE6Gphosphodiesterase 6G, cGMP-specific, rod, gamma (ENSG00000185527), score: 0.54 PDIK1LPDLIM1 interacting kinase 1 like (ENSG00000175087), score: 0.45 PFN4profilin family, member 4 (ENSG00000176732), score: 0.46 PGRprogesterone receptor (ENSG00000082175), score: 0.7 PHF12PHD finger protein 12 (ENSG00000109118), score: 0.47 PHF21APHD finger protein 21A (ENSG00000135365), score: 0.44 PHKA2phosphorylase kinase, alpha 2 (liver) (ENSG00000044446), score: 0.52 PHTF1putative homeodomain transcription factor 1 (ENSG00000116793), score: 0.74 PIGLphosphatidylinositol glycan anchor biosynthesis, class L (ENSG00000108474), score: 0.5 PIGSphosphatidylinositol glycan anchor biosynthesis, class S (ENSG00000087111), score: 0.48 PITPNC1phosphatidylinositol transfer protein, cytoplasmic 1 (ENSG00000154217), score: -0.47 PLS3plastin 3 (ENSG00000102024), score: -0.48 PPA2pyrophosphatase (inorganic) 2 (ENSG00000138777), score: -0.65 PPIP5K2diphosphoinositol pentakisphosphate kinase 2 (ENSG00000145725), score: 0.43 PPP1R8protein phosphatase 1, regulatory (inhibitor) subunit 8 (ENSG00000117751), score: 0.77 PRCPprolylcarboxypeptidase (angiotensinase C) (ENSG00000137509), score: -0.55 PRKACBprotein kinase, cAMP-dependent, catalytic, beta (ENSG00000142875), score: -0.57 PRKD1protein kinase D1 (ENSG00000184304), score: 0.55 PRSSL1protease, serine-like 1 (ENSG00000185198), score: 0.73 PSMA1proteasome (prosome, macropain) subunit, alpha type, 1 (ENSG00000129084), score: -0.44 PSMB4proteasome (prosome, macropain) subunit, beta type, 4 (ENSG00000159377), score: -0.51 PSMG1proteasome (prosome, macropain) assembly chaperone 1 (ENSG00000183527), score: 0.45 PTK2PTK2 protein tyrosine kinase 2 (ENSG00000169398), score: -0.43 PTPN12protein tyrosine phosphatase, non-receptor type 12 (ENSG00000127947), score: -0.43 PUS10pseudouridylate synthase 10 (ENSG00000162927), score: 0.53 PUS3pseudouridylate synthase 3 (ENSG00000110060), score: 0.59 PYCRLpyrroline-5-carboxylate reductase-like (ENSG00000104524), score: 0.77 RAB27ARAB27A, member RAS oncogene family (ENSG00000069974), score: -0.43 RAB38RAB38, member RAS oncogene family (ENSG00000123892), score: 0.55 RAB7L1RAB7, member RAS oncogene family-like 1 (ENSG00000117280), score: -0.43 RABEPKRab9 effector protein with kelch motifs (ENSG00000136933), score: -0.62 RABL3RAB, member of RAS oncogene family-like 3 (ENSG00000144840), score: 0.44 RBM26RNA binding motif protein 26 (ENSG00000139746), score: 0.45 RFX3regulatory factor X, 3 (influences HLA class II expression) (ENSG00000080298), score: 0.44 RG9MTD1RNA (guanine-9-) methyltransferase domain containing 1 (ENSG00000174173), score: -0.44 RGP1RGP1 retrograde golgi transport homolog (S. cerevisiae) (ENSG00000107185), score: 0.81 RIBC2RIB43A domain with coiled-coils 2 (ENSG00000128408), score: 0.48 RIC8Bresistance to inhibitors of cholinesterase 8 homolog B (C. elegans) (ENSG00000111785), score: 0.6 RNF34ring finger protein 34 (ENSG00000170633), score: 0.43 RNMTL1RNA methyltransferase like 1 (ENSG00000171861), score: -0.46 RPE65retinal pigment epithelium-specific protein 65kDa (ENSG00000116745), score: 0.66 RPGRIP1LRPGRIP1-like (ENSG00000103494), score: 0.61 RPS6KA5ribosomal protein S6 kinase, 90kDa, polypeptide 5 (ENSG00000100784), score: 0.48 RPS6KA6ribosomal protein S6 kinase, 90kDa, polypeptide 6 (ENSG00000072133), score: 0.5 RPUSD2RNA pseudouridylate synthase domain containing 2 (ENSG00000166133), score: -0.5 SAP30LSAP30-like (ENSG00000164576), score: 0.48 SARSseryl-tRNA synthetase (ENSG00000031698), score: -0.45 SCML2sex comb on midleg-like 2 (Drosophila) (ENSG00000102098), score: 0.67 SCRN2secernin 2 (ENSG00000141295), score: -0.45 SEMA3Dsema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3D (ENSG00000153993), score: 0.46 SERPINB12serpin peptidase inhibitor, clade B (ovalbumin), member 12 (ENSG00000166634), score: 0.5 SFRS8splicing factor, arginine/serine-rich 8 (suppressor-of-white-apricot homolog, Drosophila) (ENSG00000061936), score: 0.46 SGCEsarcoglycan, epsilon (ENSG00000127990), score: 0.53 SGOL1shugoshin-like 1 (S. pombe) (ENSG00000129810), score: 0.63 SH2D3CSH2 domain containing 3C (ENSG00000095370), score: -0.47 SH3YL1SH3 domain containing, Ysc84-like 1 (S. cerevisiae) (ENSG00000035115), score: -0.47 SIDT1SID1 transmembrane family, member 1 (ENSG00000072858), score: 0.51 SIK2salt-inducible kinase 2 (ENSG00000170145), score: -0.48 SKILSKI-like oncogene (ENSG00000136603), score: 0.52 SLC35B4solute carrier family 35, member B4 (ENSG00000205060), score: 0.44 SLC38A8solute carrier family 38, member 8 (ENSG00000166558), score: 0.68 SLC3A1solute carrier family 3 (cystine, dibasic and neutral amino acid transporters, activator of cystine, dibasic and neutral amino acid transport), member 1 (ENSG00000138079), score: -0.44 SLC4A7solute carrier family 4, sodium bicarbonate cotransporter, member 7 (ENSG00000033867), score: 0.64 SMARCAD1SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1 (ENSG00000163104), score: 0.48 SMC1Bstructural maintenance of chromosomes 1B (ENSG00000077935), score: 0.5 SMG7Smg-7 homolog, nonsense mediated mRNA decay factor (C. elegans) (ENSG00000116698), score: 0.45 SMUG1single-strand-selective monofunctional uracil-DNA glycosylase 1 (ENSG00000123415), score: -0.51 SNRKSNF related kinase (ENSG00000163788), score: 0.67 SNX13sorting nexin 13 (ENSG00000071189), score: 0.52 SPG20spastic paraplegia 20 (Troyer syndrome) (ENSG00000133104), score: -0.51 SRMSsrc-related kinase lacking C-terminal regulatory tyrosine and N-terminal myristylation sites (ENSG00000125508), score: 0.48 SRPX2sushi-repeat-containing protein, X-linked 2 (ENSG00000102359), score: 0.51 SSBP1single-stranded DNA binding protein 1 (ENSG00000106028), score: -0.45 ST6GALNAC4ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 4 (ENSG00000136840), score: 0.51 STACSH3 and cysteine rich domain (ENSG00000144681), score: 0.44 STAU2staufen, RNA binding protein, homolog 2 (Drosophila) (ENSG00000040341), score: -0.44 STILSCL/TAL1 interrupting locus (ENSG00000123473), score: 0.77 STK10serine/threonine kinase 10 (ENSG00000072786), score: -0.42 STRBPspermatid perinuclear RNA binding protein (ENSG00000165209), score: 0.43 STT3BSTT3, subunit of the oligosaccharyltransferase complex, homolog B (S. cerevisiae) (ENSG00000163527), score: 0.49 STX8syntaxin 8 (ENSG00000170310), score: -0.77 TAB2TGF-beta activated kinase 1/MAP3K7 binding protein 2 (ENSG00000055208), score: -0.44 TAF4BTAF4b RNA polymerase II, TATA box binding protein (TBP)-associated factor, 105kDa (ENSG00000141384), score: 0.51 TARSthreonyl-tRNA synthetase (ENSG00000113407), score: -0.45 TDRD5tudor domain containing 5 (ENSG00000162782), score: 0.5 TDRKHtudor and KH domain containing (ENSG00000182134), score: 0.47 TECTBtectorin beta (ENSG00000119913), score: 0.52 TERTtelomerase reverse transcriptase (ENSG00000164362), score: 0.9 TEX9testis expressed 9 (ENSG00000151575), score: 0.56 TGM2transglutaminase 2 (C polypeptide, protein-glutamine-gamma-glutamyltransferase) (ENSG00000198959), score: -0.42 THEMISthymocyte selection associated (ENSG00000172673), score: 0.58 THYN1thymocyte nuclear protein 1 (ENSG00000151500), score: -0.69 TM2D2TM2 domain containing 2 (ENSG00000169490), score: -0.48 TMEM100transmembrane protein 100 (ENSG00000166292), score: 0.53 TMEM138transmembrane protein 138 (ENSG00000149483), score: 0.49 TMEM18transmembrane protein 18 (ENSG00000151353), score: -0.54 TMPRSS4transmembrane protease, serine 4 (ENSG00000137648), score: 0.56 TMPRSS7transmembrane protease, serine 7 (ENSG00000176040), score: 0.65 TNKStankyrase, TRF1-interacting ankyrin-related ADP-ribose polymerase (ENSG00000173273), score: 0.54 TOE1target of EGR1, member 1 (nuclear) (ENSG00000132773), score: 0.79 TPMTthiopurine S-methyltransferase (ENSG00000137364), score: -0.59 TRAF3IP1TNF receptor-associated factor 3 interacting protein 1 (ENSG00000204104), score: 0.51 TRAF3IP3TRAF3 interacting protein 3 (ENSG00000009790), score: 0.54 TRIM35tripartite motif-containing 35 (ENSG00000104228), score: -0.65 TRIP12thyroid hormone receptor interactor 12 (ENSG00000153827), score: 0.5 TRPV2transient receptor potential cation channel, subfamily V, member 2 (ENSG00000187688), score: 0.48 TSEN54tRNA splicing endonuclease 54 homolog (S. cerevisiae) (ENSG00000182173), score: -0.46 TSNtranslin (ENSG00000211460), score: -0.57 TTLL1tubulin tyrosine ligase-like family, member 1 (ENSG00000100271), score: 0.48 TTLL9tubulin tyrosine ligase-like family, member 9 (ENSG00000131044), score: 0.53 TUFT1tuftelin 1 (ENSG00000143367), score: 0.67 TUSC3tumor suppressor candidate 3 (ENSG00000104723), score: 0.46 UAP1L1UDP-N-acteylglucosamine pyrophosphorylase 1-like 1 (ENSG00000197355), score: -0.46 UBE2G1ubiquitin-conjugating enzyme E2G 1 (UBC7 homolog, yeast) (ENSG00000132388), score: -0.44 UBTD2ubiquitin domain containing 2 (ENSG00000168246), score: 0.48 UHRF1ubiquitin-like with PHD and ring finger domains 1 (ENSG00000034063), score: 0.49 UNC45Aunc-45 homolog A (C. elegans) (ENSG00000140553), score: -0.49 USP14ubiquitin specific peptidase 14 (tRNA-guanine transglycosylase) (ENSG00000101557), score: 0.56 USP19ubiquitin specific peptidase 19 (ENSG00000172046), score: 0.51 USP49ubiquitin specific peptidase 49 (ENSG00000164663), score: 0.47 USP53ubiquitin specific peptidase 53 (ENSG00000145390), score: 0.44 UTP20UTP20, small subunit (SSU) processome component, homolog (yeast) (ENSG00000120800), score: 0.7 VWA5B1von Willebrand factor A domain containing 5B1 (ENSG00000158816), score: 0.53 WDHD1WD repeat and HMG-box DNA binding protein 1 (ENSG00000198554), score: 0.44 WDR48WD repeat domain 48 (ENSG00000114742), score: 0.75 WDR64WD repeat domain 64 (ENSG00000162843), score: 0.46 WDR77WD repeat domain 77 (ENSG00000116455), score: 0.59 XPCxeroderma pigmentosum, complementation group C (ENSG00000154767), score: -0.46 XPO4exportin 4 (ENSG00000132953), score: 0.49 ZC3H15zinc finger CCCH-type containing 15 (ENSG00000065548), score: -0.74 ZFYVE1zinc finger, FYVE domain containing 1 (ENSG00000165861), score: 0.51 ZFYVE21zinc finger, FYVE domain containing 21 (ENSG00000100711), score: -0.57 ZMYND10zinc finger, MYND-type containing 10 (ENSG00000004838), score: 0.5 ZMYND19zinc finger, MYND-type containing 19 (ENSG00000165724), score: 0.54 ZNF512zinc finger protein 512 (ENSG00000243943), score: 0.85 ZNF704zinc finger protein 704 (ENSG00000164684), score: -0.46 ZNF800zinc finger protein 800 (ENSG00000048405), score: 0.56 ZNFX1zinc finger, NFX1-type containing 1 (ENSG00000124201), score: 0.48 ZWILCHZwilch, kinetochore associated, homolog (Drosophila) (ENSG00000174442), score: 0.65
| Id | species | tissue | sex | individual |
|---|---|---|---|---|
| mdo_br_f_ca1 | mdo | br | f | _ |
| mdo_br_m_ca1 | mdo | br | m | _ |
| mdo_cb_f_ca1 | mdo | cb | f | _ |
| mdo_cb_m_ca1 | mdo | cb | m | _ |
| mdo_ts_m1_ca1 | mdo | ts | m | 1 |
| mdo_ts_m2_ca1 | mdo | ts | m | 2 |