Difference between revisions of "ExpressionView"
| (64 intermediate revisions by 4 users not shown) | |||
| Line 1: | Line 1: | ||
| − | [[ | + | [[Category:Bulletins]] |
| + | <newstitle>ExpressionView application note</newstitle> | ||
| + | <teaser> | ||
| + | We published <a href="http://bioinformatics.oxfordjournals.org/cgi/content/abstract/btq334">an application note</a> about the ExpressionView bicluster visualization tool in Bioinformatics. Please see the [[ExpressionView]] page for more — documentation, downloads, screenshots — on ExpressionView. | ||
| + | <date>29 Jul 2010 — 13:32</date> | ||
| + | </teaser> | ||
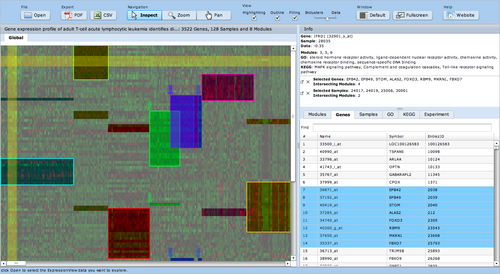
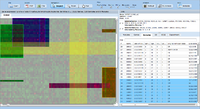
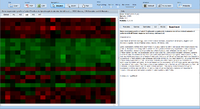
| − | ExpressionView is an R package that provides an interactive environment to explore biclusters identified in gene expression data. A sophisticated ordering algorithm is used to present the biclusters in a visually appealing layout | + | [[image:Expressionview.screenshot.png|Screenshot of the ExpressionView applet|500px|left]] |
| + | <br style="clear: both" /> | ||
| + | <br> | ||
| + | ExpressionView is an R package that provides an interactive environment to explore biclusters identified in gene expression data. A sophisticated ordering algorithm is used to present the biclusters in a visually appealing layout. From this overview, the user can select individual biclusters and access all the biologically relevant data associated with it. The package is aimed to facilitate the collaboration between bioinformaticians and life scientists who are not familiar with the R language. | ||
| − | = | + | = Demos = |
| − | * [http:// | + | |
| − | * [http:// | + | * [http://www2.unil.ch/cbg/software/expressionview/flash/ExpressionView.html?filename=../data/Expressionview.sampledata.all.small.evf Launch ExpressionView with adult T-cell acute lymphocytic leukemia (ALL) data (8 modules)] |
| − | * [http:// | + | * [http://www2.unil.ch/cbg/software/expressionview/flash/ExpressionView.html?filename=../data/Expressionview.sampledata.all.large.evf Launch ExpressionView with adult T-cell acute lymphocytic leukemia (ALL) data (108 modules)] |
| + | * [http://www2.unil.ch/cbg/software/expressionview/flash/ExpressionView.html Launch ExpressionView] | ||
| + | |||
| + | = Requirements and installation = | ||
| + | |||
| + | == Download the R package (includes the Flash applet) == | ||
| + | |||
| + | To use the ExpressionView R package you will need a working [http://www.r-project.org GNU R] installation. | ||
| + | |||
| + | As of the 23rd of April, 2010, the ExpressionView package is an official [http://www.bioconductor.org BioConductor] package. | ||
| + | |||
| + | ExpressionView depends on a number of other R packages: isa2, Biobase, AnnotationDbi, etc. The good news is that all these dependencies are installed automatically, and all you need to do is to start R and type in | ||
| + | |||
| + | source("http://bioconductor.org/biocLite.R") | ||
| + | biocLite("ExpressionView") | ||
| + | |||
| + | at your R prompt. See [http://bioconductor.org/packages/2.8/bioc/html/ExpressionView.html the ExpressionView package page at the BioConductor website] for details. | ||
| + | |||
| + | Alternatively, you can also download the package from here: | ||
| + | |||
| + | * '''[http://www.unil.ch/cbg/homepage/downloads/ExpressionView_1.0.0.zip Microsoft Windows (32 bit)]''' <br/>Download [http://www.unil.ch/cbg/homepage/downloads/ExpressionView_1.0.0.zip this file], save it in a temporary directory, and then start R. From the Packages menu choose '<code>Install packages from local zip files</code>' and select the saved file. | ||
| + | * '''[http://www.unil.ch/cbg/homepage/downloads/win64/ExpressionView_1.0.0.zip Microsoft Windows (64 bit)]''' <br/>Download [http://www.unil.ch/cbg/homepage/downloads/win64/ExpressionView_1.0.0.zip this file], save it in a temporary directory, and then start R. From the Packages menu choose '<code>Install packages from local zip files</code>' and select the saved file. | ||
| + | * '''[http://www.unil.ch/cbg/homepage/downloads/ExpressionView_1.0.0.tgz Mac OSX (Leopard)]''' <br/> Download and install [http://www.unil.ch/cbg/homepage/downloads/ExpressionView_1.0.0.tgz this file]. | ||
| + | * '''[http://www.unil.ch/cbg/homepage/downloads/ExpressionView_1.0.0.tar.gz Linux and Unix systems, R source package]''' <br/> Download [http://www.unil.ch/cbg/homepage/downloads/ExpressionView_1.0.0.tar.gz this file], save it in a temporary directory, and start R. Install the downloaded package using the <code>install.packages()</code> function: give the full path of the saved file and use the '<code>repos=NULL</code>' argument of <code>install.packages()</code>. | ||
| + | |||
| + | The Flash applet requires a Flash-enabled web browser. Please install Adobe Flash Player from the [http://get.adobe.com/flashplayer/ Adobe web site] if your browser does not have it yet. | ||
| + | |||
| + | == Download the stand-alone viewer (Adobe AIR) == | ||
| + | If you prefer a stand-alone viewer, you can download and install the Adobe AIR build [http://www2.unil.ch/cbg/software/expressionview/air/ExpressionView.air ExpressionView.air] (right-click to download file).<br> | ||
| + | To run the program, you need the AIR runtime environment which you can get from [http://get.adobe.com/air Adobe].<br> | ||
| + | ExpressionView creates files associations to .evf files, allowing you to simply double-click on such files to launch the viewer and load the data. | ||
| + | |||
| + | == Download sample data == | ||
| + | * [http://www2.unil.ch/cbg/software/expressionview/data/Expressionview.sampledata.all.small.evf Gene expression profile of adult T-cell acute lymphocytic leukemia (ALL) with 8 modules] (right-click to download file). | ||
| + | * [http://www2.unil.ch/cbg/software/expressionview/data/Expressionview.sampledata.all.large.evf Gene expression profile of adult T-cell acute lymphocytic leukemia (ALL) with 108 modules] (right-click to download file). | ||
| + | |||
| + | = License = | ||
| + | |||
| + | The ExpressionView package is licensed under the GNU General Public License, version 2 or later. For details, see http://www.gnu.org/licenses/old-licenses/gpl-2.0.html. | ||
= Screenshots = | = Screenshots = | ||
<gallery widths="200px" heights="109px" perrow="3"> | <gallery widths="200px" heights="109px" perrow="3"> | ||
| − | image:Expressionview.screenshot.1.png| | + | image:Expressionview.screenshot.1.png|Startup screen |
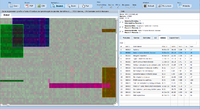
| − | image:Expressionview.screenshot.2.png| | + | image:Expressionview.screenshot.2.png|Global view after loading dataset |
| − | image:Expressionview.screenshot.3.png| | + | image:Expressionview.screenshot.3.png|Highlighting modules |
| − | image:Expressionview.screenshot.4.png| | + | image:Expressionview.screenshot.4.png|Zoom functions |
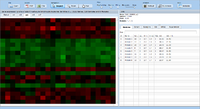
| − | image:Expressionview.screenshot.5.png| | + | image:Expressionview.screenshot.5.png|Highlighting genes (probes) and samples |
| − | image:Expressionview.screenshot.6.png| | + | image:Expressionview.screenshot.6.png|GO and KEGG associations |
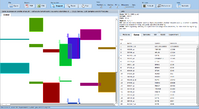
| − | image:Expressionview.screenshot.7.png| | + | image:Expressionview.screenshot.7.png|Modules view emphasizing the underlying gene expression data |
| − | image:Expressionview.screenshot.8.png| | + | image:Expressionview.screenshot.8.png|Global view without gene expression data |
| − | image:Expressionview.screenshot.9.png| | + | image:Expressionview.screenshot.9.png|Experiment description |
</gallery> | </gallery> | ||
| + | = Tutorials = | ||
| + | There are several tutorials describing how to use ExpressionView. The features of the R package are documented within the program. Just have a look at the ExpressionView help page after you have installed the package. Below, you can download the tutorial presenting the basic workflow and the description of the ordering algorithm. For the Flash applet, we have produced a few videos showing you how to use the program. | ||
| + | |||
| + | ===[[Getting started with ExpressionView|Getting started with ExpressionView]]=== | ||
| + | [[Getting started with ExpressionView|HTML]] | ||
| + | [http://www2.unil.ch/cbg/homepage/downloads/ExpressionView.tutorial.pdf PDF] | ||
| + | [http://www2.unil.ch/cbg/homepage/downloads/ExpressionView.tutorial.Rnw Rnw] | ||
| + | [http://www2.unil.ch/cbg/homepage/downloads/ExpressionView.tutorial.R R code] | ||
| − | = | + | ===[[Ordering algorithm used in ExpressionView|Ordering algorithm used in ExpressionView]]=== |
| + | [[Ordering algorithm used in ExpressionView|HTML]] | ||
| + | [http://www2.unil.ch/cbg/homepage/downloads/ExpressionView.ordering.pdf PDF] | ||
| + | ===[[ExpressionView File Format|ExpressionView File Format]]=== | ||
| + | [[ExpressionView File Format|HTML]] | ||
| + | [http://www2.unil.ch/cbg/homepage/downloads/ExpressionView.format.pdf PDF] | ||
| − | = | + | ===[http://www2.unil.ch/cbg/software/expressionview/r/ExpressionView.pdf ExpressionView R package manual]=== |
| − | + | [http://www2.unil.ch/cbg/software/expressionview/r/ExpressionView.pdf PDF] | |
| + | |||
| + | === Flash applet === | ||
| + | * [[Media:Expressionview.quickhelp.pdf|Quick help (pdf)]] | ||
| + | |||
| + | * [http://www2.unil.ch/cbg/software/expressionview/videos/Expressionview.videotutorial.getting.started.mov Getting started (video tutorial, 6 minutes)] | ||
| + | * [http://www2.unil.ch/cbg/software/expressionview/videos/Expressionview.videotutorial.tables.mov Using the tables (video tutorial, 6 minutes)] | ||
| + | * [http://www2.unil.ch/cbg/software/expressionview/videos/Expressionview.videotutorial.modularview.mov Modular view (video tutorial, 4 minutes)] | ||
| + | * [http://www2.unil.ch/cbg/software/expressionview/videos/Expressionview.videotutorial.view.mov Fullscreen feature (video tutorial, 1 minute)] | ||
| + | |||
| + | === Installing the stand-alone version === | ||
| + | * [http://www2.unil.ch/cbg/software/expressionview/videos/Expressionview.videotutorial.standalone.mov Stand-alone installation (video tutorial, 2 minutes)] | ||
| − | == | + | = Additional documentation and downloads = |
| + | The ExpressionView data file is an XML file. We have created a corresponding XML Schema file that defines its structure. | ||
| + | * [http://www2.unil.ch/cbg/software/expressionview/data/expressionview.xsd ExpressionView XML data file schema] | ||
| − | + | The Flash applet is written in ActionScript. It is open source and can be built from the command line using the Adobe Flex SDK or more conveniently with the Adobe Flex Builder IDE. For more information, visit the [http://www.adobe.com/flex Flex website]. | |
| + | * [http://www2.unil.ch/cbg/software/expressionview/source/ExpressionView.tar.gz ExpressionView source code (for the Flash applet)] | ||
| + | * [http://www2.unil.ch/cbg/software/expressionview/doc ActionScript source code documentation] | ||
| − | + | For the Flash applet, we have implemented components that could also be used in other applications. The most important one being the LargeBitmapData class that allows one to work with BitmapData of arbitrary size. In the [http://livedocs.adobe.com/flex/3/langref/flash/display/BitmapData.html standard BitmapData class], the maximum size for a BitmapData object is 8,192 pixels in width or height, and the total number of pixels cannot exceed 16,777,216 pixels. Note that the ResizablePanel class is no longer used in ExpressionView. | |
| − | + | * [[Media:Expressionview.largebitmapdata.tar.gz|ActionScript implementation of the LargeBitmapData class (allows to use bitmaps of arbitrary dimensions)]] | |
| − | * | + | * [[Media:Expressionview.resizablepanel.tar.gz|ActionScript implementation of the ResizablePanel class (a panel with open, maximize, minimize, close and resize buttons)]] |
Latest revision as of 16:07, 20 August 2012
ExpressionView is an R package that provides an interactive environment to explore biclusters identified in gene expression data. A sophisticated ordering algorithm is used to present the biclusters in a visually appealing layout. From this overview, the user can select individual biclusters and access all the biologically relevant data associated with it. The package is aimed to facilitate the collaboration between bioinformaticians and life scientists who are not familiar with the R language.
Contents
Demos
- Launch ExpressionView with adult T-cell acute lymphocytic leukemia (ALL) data (8 modules)
- Launch ExpressionView with adult T-cell acute lymphocytic leukemia (ALL) data (108 modules)
- Launch ExpressionView
Requirements and installation
Download the R package (includes the Flash applet)
To use the ExpressionView R package you will need a working GNU R installation.
As of the 23rd of April, 2010, the ExpressionView package is an official BioConductor package.
ExpressionView depends on a number of other R packages: isa2, Biobase, AnnotationDbi, etc. The good news is that all these dependencies are installed automatically, and all you need to do is to start R and type in
source("http://bioconductor.org/biocLite.R")
biocLite("ExpressionView")
at your R prompt. See the ExpressionView package page at the BioConductor website for details.
Alternatively, you can also download the package from here:
- Microsoft Windows (32 bit)
Download this file, save it in a temporary directory, and then start R. From the Packages menu choose 'Install packages from local zip files' and select the saved file. - Microsoft Windows (64 bit)
Download this file, save it in a temporary directory, and then start R. From the Packages menu choose 'Install packages from local zip files' and select the saved file. - Mac OSX (Leopard)
Download and install this file. - Linux and Unix systems, R source package
Download this file, save it in a temporary directory, and start R. Install the downloaded package using theinstall.packages()function: give the full path of the saved file and use the 'repos=NULL' argument ofinstall.packages().
The Flash applet requires a Flash-enabled web browser. Please install Adobe Flash Player from the Adobe web site if your browser does not have it yet.
Download the stand-alone viewer (Adobe AIR)
If you prefer a stand-alone viewer, you can download and install the Adobe AIR build ExpressionView.air (right-click to download file).
To run the program, you need the AIR runtime environment which you can get from Adobe.
ExpressionView creates files associations to .evf files, allowing you to simply double-click on such files to launch the viewer and load the data.
Download sample data
- Gene expression profile of adult T-cell acute lymphocytic leukemia (ALL) with 8 modules (right-click to download file).
- Gene expression profile of adult T-cell acute lymphocytic leukemia (ALL) with 108 modules (right-click to download file).
License
The ExpressionView package is licensed under the GNU General Public License, version 2 or later. For details, see http://www.gnu.org/licenses/old-licenses/gpl-2.0.html.
Screenshots
Tutorials
There are several tutorials describing how to use ExpressionView. The features of the R package are documented within the program. Just have a look at the ExpressionView help page after you have installed the package. Below, you can download the tutorial presenting the basic workflow and the description of the ordering algorithm. For the Flash applet, we have produced a few videos showing you how to use the program.
Getting started with ExpressionView
Ordering algorithm used in ExpressionView
ExpressionView File Format
ExpressionView R package manual
Flash applet
- Getting started (video tutorial, 6 minutes)
- Using the tables (video tutorial, 6 minutes)
- Modular view (video tutorial, 4 minutes)
- Fullscreen feature (video tutorial, 1 minute)
Installing the stand-alone version
Additional documentation and downloads
The ExpressionView data file is an XML file. We have created a corresponding XML Schema file that defines its structure.
The Flash applet is written in ActionScript. It is open source and can be built from the command line using the Adobe Flex SDK or more conveniently with the Adobe Flex Builder IDE. For more information, visit the Flex website.
For the Flash applet, we have implemented components that could also be used in other applications. The most important one being the LargeBitmapData class that allows one to work with BitmapData of arbitrary size. In the standard BitmapData class, the maximum size for a BitmapData object is 8,192 pixels in width or height, and the total number of pixels cannot exceed 16,777,216 pixels. Note that the ResizablePanel class is no longer used in ExpressionView.