Difference between revisions of "ExpressionView"
| Line 5: | Line 5: | ||
= Launch and download = | = Launch and download = | ||
| + | The ExpressionView package contains two independent parts: An ordering algorithm implemented in C++ and an Adobe Flash applet written in ActionScript and Adobe Flex. These two programs are wrapped in an R package, allowing the user to take advantage of the powerful microarray analysis tools provided by the Bioconductor project. The Flash applet itself does not require a working R environment, it can be launched in any web browser via the links provided below. If you prefer a stand-alone program, you can also download the platform independent Adobe AIR version. | ||
== Launch the ExpressionView Flash applet == | == Launch the ExpressionView Flash applet == | ||
| Line 39: | Line 40: | ||
= Tutorials = | = Tutorials = | ||
| − | There are several tutorials describing how to use ExpressionView | + | There are several tutorials describing how to use ExpressionView. The features of the R package are documented within the program. Just have a look at the ExpressionView help page after you have installed the package. Below, you can download the tutorial presenting the basic workflow and the description of the ordering algorithm. For the Flash applet, we have produced a few videos showing you how to use the program. |
== R package == | == R package == | ||
| Line 58: | Line 59: | ||
= Additional documentation and downloads = | = Additional documentation and downloads = | ||
| + | The Flash applet is written in ActionScript. It is open source and can be built on the command line with the Adobe Flex SDK or more conveniently with Adobe Flex Builder IDE. For more information, visit the [[http://www.adobe.com/flex Flex website]]. | ||
* [http://www2.unil.ch/cbg/software/expressionview/doc ActionScript source code documentation] | * [http://www2.unil.ch/cbg/software/expressionview/doc ActionScript source code documentation] | ||
* [[Media:Expressionview.xmldescription.pdf|XML file structure]] | * [[Media:Expressionview.xmldescription.pdf|XML file structure]] | ||
* [[Media:Expressionview.largebitmapdata.tar.gz|ActionScript implementation of the LargeBitmapData class (allows to use bitmaps of arbitrary dimensions)]] | * [[Media:Expressionview.largebitmapdata.tar.gz|ActionScript implementation of the LargeBitmapData class (allows to use bitmaps of arbitrary dimensions)]] | ||
* [[Media:Expressionview.resizablepanel.tar.gz|ActionScript implementation of the ResizablePanel class (a panel with open, maximize, minimize, close and resize buttons)]] | * [[Media:Expressionview.resizablepanel.tar.gz|ActionScript implementation of the ResizablePanel class (a panel with open, maximize, minimize, close and resize buttons)]] | ||
Revision as of 09:23, 3 December 2009
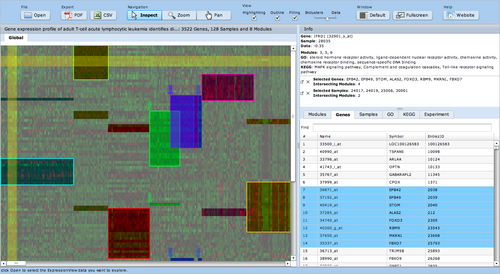
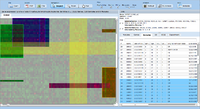
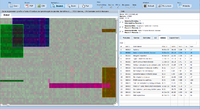
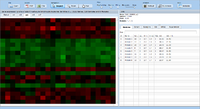
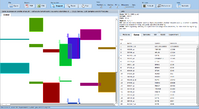
ExpressionView is an R package that provides an interactive environment to explore biclusters identified in gene expression data. A sophisticated ordering algorithm is used to present the biclusters in a visually appealing layout. From this overview, the user can select individual biclusters and access all the biologically relevant data associated with it. The package is aimed to facilitate the collaboration between bioinformaticians and life scientists who are not familiar with the R language.
Launch and download
The ExpressionView package contains two independent parts: An ordering algorithm implemented in C++ and an Adobe Flash applet written in ActionScript and Adobe Flex. These two programs are wrapped in an R package, allowing the user to take advantage of the powerful microarray analysis tools provided by the Bioconductor project. The Flash applet itself does not require a working R environment, it can be launched in any web browser via the links provided below. If you prefer a stand-alone program, you can also download the platform independent Adobe AIR version.
Launch the ExpressionView Flash applet
- Launch ExpressionView
- Launch ExpressionView with adult T-cell acute lymphocytic leukemia (ALL) data (8 modules)
- Launch ExpressionView with adult T-cell acute lymphocytic leukemia (ALL) data (108 modules)
Download the R package (includes the Flash applet)
- Windows binary package
- Mac OS X 32 bit binary package, Mac OS X 64 bit binary package
- Source package
Download the stand-alone viewer (Adobe AIR)
If you prefer a stand-alone viewer, you can download and install the Adobe AIR build ExpressionView.air (right-click to download file).
To run the program, you need the AIR runtime environment which you can get from Adobe.
ExpressionView creates files associations to .evf files, allowing you to simply double-click on such files to launch the viewer and load the data.
Download sample data
- Gene expression profile of adult T-cell acute lymphocytic leukemia (ALL) with 8 modules (right-click to download file).
- Gene expression profile of adult T-cell acute lymphocytic leukemia (ALL) with 108 modules (right-click to download file).
Screenshots
Tutorials
There are several tutorials describing how to use ExpressionView. The features of the R package are documented within the program. Just have a look at the ExpressionView help page after you have installed the package. Below, you can download the tutorial presenting the basic workflow and the description of the ordering algorithm. For the Flash applet, we have produced a few videos showing you how to use the program.
R package
Flash applet
- Getting started (video tutorial, 6 minutes)
- Using the tables (video tutorial, 6 minutes)
- Modular view (video tutoria, 4 minutesl)
- Fullscreen feature (video tutorial, 1 minute)
Installing the stand-alone version
Additional documentation and downloads
The Flash applet is written in ActionScript. It is open source and can be built on the command line with the Adobe Flex SDK or more conveniently with Adobe Flex Builder IDE. For more information, visit the [Flex website].