Difference between revisions of "HypoPhen"
| (129 intermediate revisions by 3 users not shown) | |||
| Line 1: | Line 1: | ||
| + | [[Category:Bulletins]] | ||
| + | <newstitle> CBG's high troughput plant phenotyping software HypoPhen helps understand phototropism in plants</newstitle> | ||
| + | <teaser> | ||
| + | In collaboration with the group of Christian Fankhauser at CIG, UNIL, we developed the HypoPhen software for the high throughput quantification of seedling elongation and bending from time-lapsed images. Using this tool, hundreds of Arabidopsis seedlings were measured to show that phytochrome A in the nucleus is important for phototropism. The results have been published in <a href="http://www.plantcell.org/cgi/content/short/tpc.111.095083?keytype=ref&ijkey=krNVCQ5WJMpbgHV">Plant Cell</a> on February 28 2012. | ||
| + | </teaser> | ||
| + | |||
== Introduction == | == Introduction == | ||
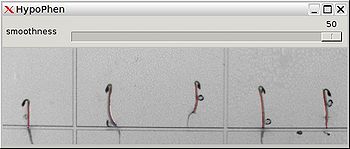
| + | Hypophen is an open-source software enabling the semi-automatic phenotyping of growing seedlings from time-lapse images. More precisely, it computes and records the elongation and bending of the seedlings. It is semi-automatic in the sense that manual calibration, verification and adjustments are sometimes needed. In my experience, it allows a throughput of about 50 images of 20 hypocotyls in about 10 minutes, given a reasonable image quality. | ||
| − | + | The software was developed within the context described in the following paper: | |
| + | <pubmed> | ||
| + | 22374392 | ||
| + | </pubmed> | ||
| + | [[Image:HypoPhen_screenshot1.jpg| thumb | screenshot | 350px]] | ||
| + | |||
| + | === Movies === | ||
| + | Here is a [[Media:HypoPhen_movie1.avi | movie]] showing in real time an excerpt from the semi-automated processing of images of 14 hypocotyls. In those 20 seconds, five frames (70 hypocotyls) are processed. Here is another [[Media:HypoPhen_movie2.avi | movie]] showing the analysis from scratch of the 12 example images of 5 hypocotyls, including the manual calibration procedure. | ||
| + | |||
| + | == Prerequisite == | ||
| + | Hypophen works on '''Linux''', '''Mac OS X''' version 10.6 or later and '''Windows'''. | ||
| + | |||
| + | For Linux, it needs the [http://opencv.willowgarage.com/wiki/ OpenCV] library (free) and must be compiled from source, The use of [http://www.cmake.org CMake] (also free) makes this rather straightforward. You also need a two button mouse to use the software. More details are given in the manual. | ||
| + | |||
| + | Windows and Mac user can directly download the executable file and launch it from the command prompt | ||
== Download == | == Download == | ||
| − | + | You can download the [[Media:Hypophen.tar.gz | C++ source code]] (version 0.4). The latest code is available on [https://sourceforge.net/p/hypophen sourceforge] | |
| + | |||
| + | The windows executable of version 0.4 (along with required dlls) for Windows 7 is [[Media:HypoPhen_win.zip | here]]. | ||
| + | Note that this executable is for the standard x86 (32 bits) architectures. Let [[User:Micha | Micha]] know if you need it for a 64 bits architecture. | ||
| + | |||
| + | There is also a [[Media:HypoPhen.dmg.zip | dmg file]] for MacOS (version 0.4) | ||
| + | |||
| + | The [[Media:HypoPhenManual.pdf | manual ]] explains how to install and use the software. | ||
| + | |||
| + | You can also download a small set of hypocotyl [[Media:HypoPhenTestImages.zip | test images]] to test the software. Those images were kindly provided by Emilie Demarsy. | ||
| + | |||
| + | == Quick testing == | ||
| + | |||
| + | For '''Mac OS''', you can download the [[Media:HypoPhen.dmg.zip | dmg file]] and the [[Media:HypoPhenTestImages.zip | test images]]. Unzip the test images and put the "images" folder in the hypoPhen.app/Contents/MacOS/ folder (yes, you have to enter the hypoPhen application folder). Double click on hypoPhen file in this same folder, will launch the software on the test images. Refer to the manual for usage and more detailed instructions. | ||
| + | |||
| + | |||
| + | For '''Windows 7''', you can download the [[Media:HypoPhen_win.zip | windows executable]] and the [[Media:HypoPhenTestImages.zip | test images]]. Unzip both files and put the "images" folder in the "HypoPhen_win" folder. Double click on hypoPhen file in this same folder, will launch the software on the test images. Refer to the manual for usage and more detailed instructions. | ||
| + | |||
| + | == Related software == | ||
| + | Software trying to achieve similar goals include [http://brie.cshl.edu/~liyawang/HYPOTrace HypoTrace] and [http://cactus.salk.edu/hyde/ HyDe] both of which are matlab-based and not open-source. | ||
| + | |||
| + | == Benefiting and contributing == | ||
| + | This software is provided "as is", in the hope that it will be useful but without any warranty of any kind. If you use this software for research purposes, please be kind enough to mention it in your scientific publications by citing the [http://www.plantcell.org/cgi/content/short/tpc.111.095083?keytype=ref&ijkey=krNVCQ5WJMpbgHV Plant Cell paper] above. If you find any bug, have any problem with the installation or would like to contribute to further developing this software, please write an email to [[User:Micha|Micha Hersch]]. | ||
| + | The project is also available on [https://sourceforge.net/p/hypophen sourceforge] | ||
== Credits == | == Credits == | ||
| − | Hypophen was written by [[User:Micha|Micha Hersch]] in collaboration with Chitose Kami and Christian Fankhauser from the Center for Integrative Genomics at the University of Lausanne. The project was initiated with the help of Ioannis Xenarios from Vital-IT and Sven Bergmann, head of the CBG. It uses the OpenCV library and some code written by Basilio Noris. | + | Hypophen was written by [[User:Micha|Micha Hersch]] in collaboration with Chitose Kami and Christian Fankhauser from the Center for Integrative Genomics at the University of Lausanne. The project was initiated with the help of Ioannis Xenarios from Vital-IT and [[User:Sven|Sven Bergmann]], head of the CBG. It uses the OpenCV library and some code written by Basilio Noris. Emilie Demarsy provided useful feedback and example of images. |
| + | |||
| + | The development of this software was funded by [http://www.systemsx.ch SystemsX] throught the [https://wiki.systemsx.ch/display/PGRTDproj/Home Plant Growth] project | ||
| + | |||
| + | [[Image:Plantgrowthlogo.png| plant growth| thumb |100 px]] | ||
| + | [[File:SystemsXlogo.png| sytemsX |250 px]] | ||
| + | |||
| + | ---- | ||
Latest revision as of 13:09, 14 February 2017
Contents
Introduction
Hypophen is an open-source software enabling the semi-automatic phenotyping of growing seedlings from time-lapse images. More precisely, it computes and records the elongation and bending of the seedlings. It is semi-automatic in the sense that manual calibration, verification and adjustments are sometimes needed. In my experience, it allows a throughput of about 50 images of 20 hypocotyls in about 10 minutes, given a reasonable image quality.
The software was developed within the context described in the following paper:
Kami C, Hersch M, Trevisan M, Genoud T, Hiltbrunner A, Bergmann S, Fankhauser C
Nuclear phytochrome A signaling promotes phototropism in Arabidopsis.
Plant Cell: 2012 Feb, 24(2);566-76
[PubMed:22374392]
[WorldCat.org:
ISSN
ESSN
]
[DOI]
( o)
Movies
Here is a movie showing in real time an excerpt from the semi-automated processing of images of 14 hypocotyls. In those 20 seconds, five frames (70 hypocotyls) are processed. Here is another movie showing the analysis from scratch of the 12 example images of 5 hypocotyls, including the manual calibration procedure.
Prerequisite
Hypophen works on Linux, Mac OS X version 10.6 or later and Windows.
For Linux, it needs the OpenCV library (free) and must be compiled from source, The use of CMake (also free) makes this rather straightforward. You also need a two button mouse to use the software. More details are given in the manual.
Windows and Mac user can directly download the executable file and launch it from the command prompt
Download
You can download the C++ source code (version 0.4). The latest code is available on sourceforge
The windows executable of version 0.4 (along with required dlls) for Windows 7 is here. Note that this executable is for the standard x86 (32 bits) architectures. Let Micha know if you need it for a 64 bits architecture.
There is also a dmg file for MacOS (version 0.4)
The manual explains how to install and use the software.
You can also download a small set of hypocotyl test images to test the software. Those images were kindly provided by Emilie Demarsy.
Quick testing
For Mac OS, you can download the dmg file and the test images. Unzip the test images and put the "images" folder in the hypoPhen.app/Contents/MacOS/ folder (yes, you have to enter the hypoPhen application folder). Double click on hypoPhen file in this same folder, will launch the software on the test images. Refer to the manual for usage and more detailed instructions.
For Windows 7, you can download the windows executable and the test images. Unzip both files and put the "images" folder in the "HypoPhen_win" folder. Double click on hypoPhen file in this same folder, will launch the software on the test images. Refer to the manual for usage and more detailed instructions.
Related software
Software trying to achieve similar goals include HypoTrace and HyDe both of which are matlab-based and not open-source.
Benefiting and contributing
This software is provided "as is", in the hope that it will be useful but without any warranty of any kind. If you use this software for research purposes, please be kind enough to mention it in your scientific publications by citing the Plant Cell paper above. If you find any bug, have any problem with the installation or would like to contribute to further developing this software, please write an email to Micha Hersch. The project is also available on sourceforge
Credits
Hypophen was written by Micha Hersch in collaboration with Chitose Kami and Christian Fankhauser from the Center for Integrative Genomics at the University of Lausanne. The project was initiated with the help of Ioannis Xenarios from Vital-IT and Sven Bergmann, head of the CBG. It uses the OpenCV library and some code written by Basilio Noris. Emilie Demarsy provided useful feedback and example of images.
The development of this software was funded by SystemsX throught the Plant Growth project