Difference between revisions of "Pascal"
| (6 intermediate revisions by 3 users not shown) | |||
| Line 4: | Line 4: | ||
We recently published Pascal (Pathway scoring algorithm), a tool that allows gene and pathway-level analysis of GWAS association results without the need to access the original genotypic data. Pascal was designed to be fast, accurate and to have high power to detect relevant pathways. We extensively tested our | We recently published Pascal (Pathway scoring algorithm), a tool that allows gene and pathway-level analysis of GWAS association results without the need to access the original genotypic data. Pascal was designed to be fast, accurate and to have high power to detect relevant pathways. We extensively tested our | ||
approach on a large collection of real GWAS association results and saw better discovery of confirmed pathways than with other popular methods. The paper is available in <a href=http://journals.plos.org/ploscompbiol/article?id=10.1371/journal.pcbi.1004714 > Plos Computational Biology </a>. | approach on a large collection of real GWAS association results and saw better discovery of confirmed pathways than with other popular methods. The paper is available in <a href=http://journals.plos.org/ploscompbiol/article?id=10.1371/journal.pcbi.1004714 > Plos Computational Biology </a>. | ||
| − | <date> Jan | + | <date> 25 Jan 2017 </date> |
</teaser> | </teaser> | ||
| − | |||
== Rigorous gene and pathway analysis of GWAS == | == Rigorous gene and pathway analysis of GWAS == | ||
'''Pascal (Pathway scoring algorithm) is an easy-to-use tool for gene scoring and pathway analysis from GWAS results'''. Pascal uses external data to estimate linkage disequilibrium. Therefore, the user only needs to supply genome wide SNP p-values. Pascal then derives p-values for genes and predefined pathways. Pascal doesn’t use Monte-Carlo simulation to derive gene p-values. This leads to increased speed and accuracy. This speed in the gene scoring is then leveraged to control the false positive rate in pathway scoring. For pathway scoring we implemented and tested enrichment strategies that compared very favorably compared to hypergeometric enrichment. This comparison was done on a large collection of GWAS results giving us confidence to recommend Pascal for downstream analysis of GWAS results. | '''Pascal (Pathway scoring algorithm) is an easy-to-use tool for gene scoring and pathway analysis from GWAS results'''. Pascal uses external data to estimate linkage disequilibrium. Therefore, the user only needs to supply genome wide SNP p-values. Pascal then derives p-values for genes and predefined pathways. Pascal doesn’t use Monte-Carlo simulation to derive gene p-values. This leads to increased speed and accuracy. This speed in the gene scoring is then leveraged to control the false positive rate in pathway scoring. For pathway scoring we implemented and tested enrichment strategies that compared very favorably compared to hypergeometric enrichment. This comparison was done on a large collection of GWAS results giving us confidence to recommend Pascal for downstream analysis of GWAS results. | ||
Pascal is mainly written in Java and has been tested on Unix systems and Mac OsX. | Pascal is mainly written in Java and has been tested on Unix systems and Mac OsX. | ||
| + | |||
| + | ''News'' | ||
| + | * The Pascal paper was among the '''top 50 most downloaded papers''' from PLoS journals in 2016. | ||
''Download'' | ''Download'' | ||
| − | * '''[http://www2.unil.ch/cbg/images/3/3d/PASCAL.zip Pascal package]''' (Download might take a while because the 1KG-EUR data | + | * '''[http://www2.unil.ch/cbg/images/3/3d/PASCAL.zip Pascal package]''' (Download might take a while because the 1KG-EUR data are included) |
| − | * '''[[PascalTestData | Test data]]''' (Additional data that | + | * '''[[PascalTestData | Test data]]''' (Additional data that were used for evaluation in the paper) |
| + | '''Note''': We found an issue with the genotype files packaged with the version of Pascal prior to June 6th 2017 (thanks to Sujoy Ghosh for pointing us to this issue). Genotypes on chromosome 1 seemed to be truncated leading to loss of gene scores of about 5% overall (other gene scores are unchanged). We now updated the genotypes files. While, the pathway scores are well calibrated in both cases, one would expect a small drop in power. We investigated this issue on a large GWAS collection showing small power gains for the updated genotype files in the investigated settings (see result [[Updated_vs_deprecated_genotypes| here]]). | ||
''Reference'' | ''Reference'' | ||
| Line 25: | Line 28: | ||
('''b''') We use external databases to define gene sets for each reported pathway. We then compute pathway scores by combining the scores of genes that belong to the same pathways, i.e. gene sets. The fast gene scoring method allows us to dynamically recalculate gene scores by aggregating SNP p-values across pathway genes that are in LD and thus cannot be treated independently. This amounts to fusing the genes and computing a new score that takes the full LD structure of the corresponding locus into account. We evaluate pathway enrichment of high-scoring (potentially fused) genes using one of two parameter-free procedures (chi-square or empirical score), avoiding any p-value cutoffs inherent to standard binary enrichment tests. | ('''b''') We use external databases to define gene sets for each reported pathway. We then compute pathway scores by combining the scores of genes that belong to the same pathways, i.e. gene sets. The fast gene scoring method allows us to dynamically recalculate gene scores by aggregating SNP p-values across pathway genes that are in LD and thus cannot be treated independently. This amounts to fusing the genes and computing a new score that takes the full LD structure of the corresponding locus into account. We evaluate pathway enrichment of high-scoring (potentially fused) genes using one of two parameter-free procedures (chi-square or empirical score), avoiding any p-value cutoffs inherent to standard binary enrichment tests. | ||
| − | <br> | + | <br><br> |
== Tissue-specific regulatory circuits disrupted in complex disease == | == Tissue-specific regulatory circuits disrupted in complex disease == | ||
Latest revision as of 13:38, 17 January 2018
Rigorous gene and pathway analysis of GWAS
Pascal (Pathway scoring algorithm) is an easy-to-use tool for gene scoring and pathway analysis from GWAS results. Pascal uses external data to estimate linkage disequilibrium. Therefore, the user only needs to supply genome wide SNP p-values. Pascal then derives p-values for genes and predefined pathways. Pascal doesn’t use Monte-Carlo simulation to derive gene p-values. This leads to increased speed and accuracy. This speed in the gene scoring is then leveraged to control the false positive rate in pathway scoring. For pathway scoring we implemented and tested enrichment strategies that compared very favorably compared to hypergeometric enrichment. This comparison was done on a large collection of GWAS results giving us confidence to recommend Pascal for downstream analysis of GWAS results. Pascal is mainly written in Java and has been tested on Unix systems and Mac OsX.
News
- The Pascal paper was among the top 50 most downloaded papers from PLoS journals in 2016.
Download
- Pascal package (Download might take a while because the 1KG-EUR data are included)
- Test data (Additional data that were used for evaluation in the paper)
Note: We found an issue with the genotype files packaged with the version of Pascal prior to June 6th 2017 (thanks to Sujoy Ghosh for pointing us to this issue). Genotypes on chromosome 1 seemed to be truncated leading to loss of gene scores of about 5% overall (other gene scores are unchanged). We now updated the genotypes files. While, the pathway scores are well calibrated in both cases, one would expect a small drop in power. We investigated this issue on a large GWAS collection showing small power gains for the updated genotype files in the investigated settings (see result here).
Reference
- Fast and rigorous computation of gene and pathway scores from SNP-based summary statistics. (PDF)
Lamparter D*, Marbach D*, Rueedi R, Kutalik Z, and Bergmann S.
PLoS Computational Biology 12, e1004714, 2016.
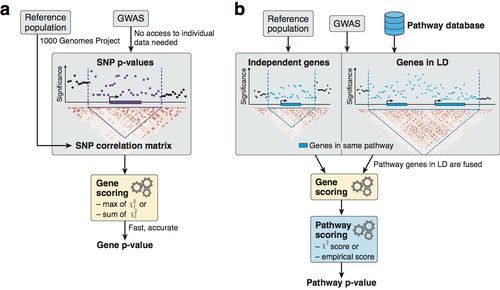
Figure: Overview of methodology to compute gene and pathway scores
(a) We compute gene scores by aggregating SNP p-values from a GWAS meta-analysis (without the need for individual genotypes), while correcting for linkage disequilibrium (LD) structure. To this end, we use numerical and analytic solutions to compute gene p-values efficiently and accurately given LD information from a reference population. Two options are available: the max and sum of chi-squared statistics, which are based on the most significant SNP and the average association signal across the region, respectively.
(b) We use external databases to define gene sets for each reported pathway. We then compute pathway scores by combining the scores of genes that belong to the same pathways, i.e. gene sets. The fast gene scoring method allows us to dynamically recalculate gene scores by aggregating SNP p-values across pathway genes that are in LD and thus cannot be treated independently. This amounts to fusing the genes and computing a new score that takes the full LD structure of the corresponding locus into account. We evaluate pathway enrichment of high-scoring (potentially fused) genes using one of two parameter-free procedures (chi-square or empirical score), avoiding any p-value cutoffs inherent to standard binary enrichment tests.
Tissue-specific regulatory circuits disrupted in complex disease
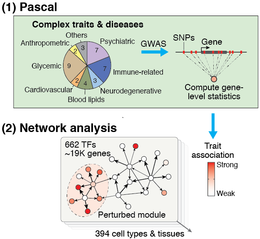
The efficiency and accuracy of Pascal opens the door to large-scale analyses that would not have been possible with previous tools. For example, summarizing SNP p-values at the level of genes is a crucial step in most network-based GWAS analysis methods. Pascal was key for our recent work, where we integrated 37 GWAS datasets with close to 400 tissue-specific gene regulatory circuits to systematically analyze the inter-connectivity of genes that are perturbed by trait-associated genetic variants. This study showed that disease-associated genetic variants often disturb regulatory modules in cell types or tissues that are highly specific to that disease, giving new insights on disease mechanisms.
Reference
- Tissue-specific regulatory circuits reveal variable modular perturbations across complex diseases. (PDF)
Marbach D, Lamparter D, Quon G, Kellis M, Kutalik Z, and Bergmann S.
Nature Methods, 13, 366-370, 2016.