Difference between revisions of "Pascal"
| Line 1: | Line 1: | ||
[[Category:Bulletins]] | [[Category:Bulletins]] | ||
| − | <newstitle> A new pathway analysis method for GWAS summary statistics</newstitle> | + | <newstitle> A new pathway analysis method and software for GWAS summary statistics</newstitle> |
<teaser> | <teaser> | ||
| − | We recently published Pascal (Pathway scoring algorithm), a tool that allows gene and pathway-level | + | We recently published Pascal (Pathway scoring algorithm), a tool that allows gene and pathway-level analysis of GWAS association results without the need to access the original genotypic data. Pascal was designed to be fast, accurate and to have high power to detect relevant pathways. We extensively tested our |
| − | + | approach on a large collection of real GWAS association results and saw better discovery of confirmed pathways than with other popular methods. The paper is available in <a href=http://journals.plos.org/ploscompbiol/article?id=10.1371/journal.pcbi.1004714 > Plos Computational Biology </a>. | |
| − | |||
| − | approach on a large collection of real GWAS association results and saw better discovery of confirmed | ||
| − | |||
<date> Jan 25 </date> | <date> Jan 25 </date> | ||
</teaser> | </teaser> | ||
Revision as of 16:20, 2 February 2016
Pascal (Pathway scoring algorithm) is an easy-to-use tool for gene scoring and pathway analysis from GWAS results. Pascal uses external data to estimate linkage disequilibrium. Therefore, the user only needs to supply genome wide SNP p-values. Pascal then derives p-values for genes and predefined pathways. Pascal doesn’t use Monte-Carlo simulation to derive gene p-values. This leads to increased speed and accuracy. This speed in the gene scoring is then leveraged to control the false positive rate in pathway scoring. For pathway scoring we implemented and tested enrichment strategies that compared very favorably compared to hypergeometric enrichment. This comparison was done on a large collection of GWAS results giving us confidence to recommend Pascal for downstream analysis of GWAS results.
Pascal is mainly written in Java and has been tested on Unix systems and Mac OsX.
You can download the package here. (Note: This might take a while because it included the 1KG-EUR data.)
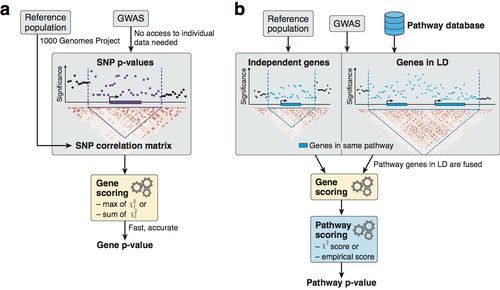
Overview of the methodology to compute gene and pathway scores.
a) We compute gene scores by aggregating SNP p-values from a GWAS meta-analysis (without the need for individual genotypes), while correcting for linkage disequilibrium (LD) structure. To this end, we use numerical and analytic solutions to compute gene p-values efficiently and accurately given LD information from a reference population. Two options are available: the max and sum of chi-squared statistics, which are based on the most significant SNP and the average association signal across the region, respectively.
b) We use external databases to define gene sets for each reported pathway. We then compute pathway scores by combining the scores of genes that belong to the same pathways, i.e. gene sets. The fast gene scoring method allows us to dynamically recalculate gene scores by aggregating SNP p-values across pathway genes that are in LD and thus cannot be treated independently. This amounts to fusing the genes and computing a new score that takes the full LD structure of the corresponding locus into account. We evaluate pathway enrichment of high-scoring (potentially fused) genes using one of two parameter-free procedures (chi-square or empirical score), avoiding any p-value cutoffs inherent to standard binary enrichment tests.
Test Data
Additional data that was used in evaluating the tool can be found here.